Sequencing technology keeps getting better, faster, more productive — but that’s not the only thing that shapes the big picture of where we’re headed as a field. If you want to understand where genomics is going, you should pay attention to the Global Alliance for Genomics and Health, or GA4GH.
GA4GH is an international organization composed largely of contributors from member institutions in healthcare, research, patient advocacy, and information technology, seeking to enable responsible genomic data sharing within a human rights framework.
The boring way to describe what GA4GH does is to say it develops standards and policies — ranging from technology standards like file formats and application programming interfaces (APIs), which aim to enable interoperability and broad access to data and tools among the global research community, to policy frameworks like patient consent clauses and data privacy policies.
I prefer to think of it this way: GA4GH contributors are effectively building the scaffolding for what genomics will deliver in practice, at scale, over the next decade.
The GA4GH standard development process involves collaboration with implementers, i.e. groups that apply the GA4GH standards in practice, typically in the context of driver projects — which have the advantage of providing real-life use cases. (As it turns out, you develop better solutions when you’re actually working on real problems.)
For technology standards, implementation means building software tools and operating services that follow the rules laid out by the relevant standards.
For example, the CRAM and VCF file formats are widely-used bioinformatics standards stewarded by GA4GH that specify how to encode sequencing reads and variant calls, respectively. There is a software library called htsjdk that implements both of these standards in the Java programming language, meaning that it includes code that is capable of reading and writing files that are encoded according to those standards. Researchers can then use genome analysis tools like GATK and Picard, which include the htsjdk library, to read and write CRAM and VCF files as part of their analysis work. Tadaa. (For fans of C, substitute htslib and samtools in the library/tool mentions.)
In addition to these now-classic (if imperfect) workhorse formats, GA4GH has been driving the development of other, newer knowledge representation standards that you may not yet be aware of, but will likely transform the way many of us work. Take the Variation Representation Specification (VRS, pronounced “verse”), which among other things makes it possible to capture the complex information that underlies variant interpretation in computable form, a key feature for solving variant interpretation bottlenecks. Right now, VRS is a fairly niche product, but within a few years I expect we’ll see it being used across a variety of research and clinical diagnostic platforms.
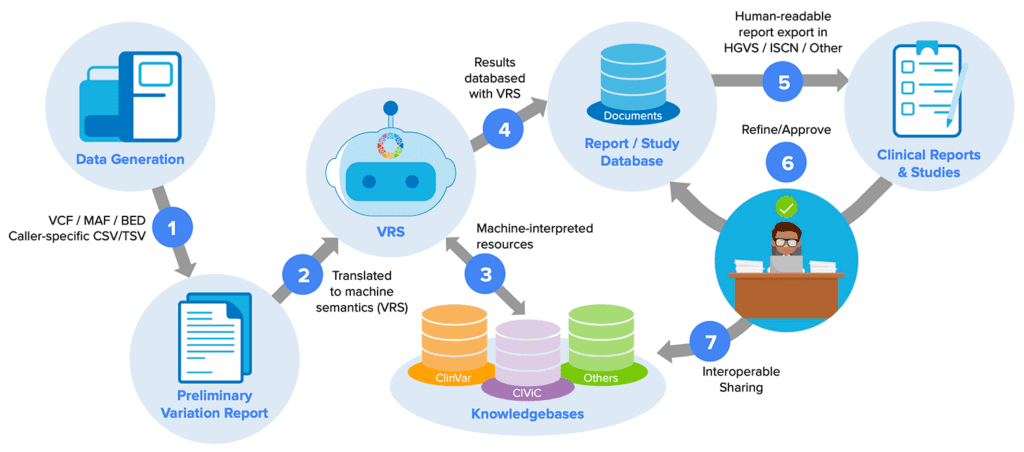

Computable standards for alleviating variant interpretation bottlenecks. From “Genomic Knowledge Standards Advancements” by Larry Babb, presented at the 10th Plenary Meeting of the GA4GH (see Day 1 recording).
Pro-tip: you can check out the Python implementation of VRS in Github, and there’s even a Terra workspace hosting Jupyter notebooks that demonstrate how to use it in practice.
Speaking of platforms, another major axis of GA4GH standard development is platform-level interoperability, i.e. infrastructure standards that enable platforms like Terra to talk to each other.
I’ve written before about the big picture of interoperability for open ecosystems, the example of the AnVIL project, and how Terra uses the DRS standard for data interoperability specifically. The ultimate goal here is to make it possible for researchers to do things like combining data from separate repositories into powerful federated analyses without having to move any of it around.
Excitingly, that dream is starting to materialize! We are now at the stage where data federation is effectively possible — for a limited set of datasets and platforms, with some clunkiness involved. As the work continues, you can expect the scope of what’s possible to include more datasets, with a smoother experience as the handoff between platforms gets ironed out.
There is a lot more to say about the scope and impact of GA4GH work, but I’ll have to leave that for another time.
The bottom line is, if any of this is new to you, now is a great time to start getting caught up.
The organization held its annual plenary meeting over two days last week, and the full recordings for both Day 1 and Day 2 are available on YouTube, annotated with timestamps for specific sessions and presentations (see the expanded video descriptions). You can also find links to the slide decks in the agenda.
As a new member of the organization (I joined the Large-Scale Genomics Work Stream in June), I found the lineup of talks struck a great balance between showcasing progress made so far, outlining upcoming challenges and discussing concrete solutions. I hope you will find these resources useful too — and consider joining the effort!
Additional resources
For a more comprehensive tour of the Global Alliance’s scope, vision, and outputs, read the “Perspective” paper published late last year in Cell Genomics by Heidi Rehm and colleagues:
GA4GH: International policies and standards for data sharing across genomic research and healthcare (2021) Cell Genomics, Volume 1, Issue 2, https://doi.org/10.1016/j.xgen.2021.100029



