We’ve name-dropped the AnVIL project a number of times in recent blog posts — covering topics such as Dockstore, RStudio, Galaxy, and of course who can forget DRS URIs — so it feels like it’s high time that we gave you some context for this beyond just sending you to the AnVIL website for more information. Specifically, we thought it might be useful to reiterate the key goals of the AnVIL project and explain briefly how Terra fits in with the other AnVIL components. Please do keep in mind that this is written from the Terra perspective, and is not intended to represent the AnVIL project in its entirety, although I’ve done my best to provide a high-level summary for your convenience.
As a prelude, the bare minimum you need to know is that AnVIL stands for Analysis, Visualization, and Informatics Lab-Space, and is an initiative driven by the National Human Genome Research Institute (NHGRI), a subdivision of the National Institutes of Health (NIH), to empower the genomics research community to use cloud infrastructure for data sharing and computing. This initiative is supported by two distinct NHGRI awards, one led by the Broad Institute in Cambridge and the other led by Johns Hopkins University in Baltimore, pulling together contributors from a dozen institutions across the USA.

Project sites participating in the AnVIL project (source: NHGRI Funded Programs website)
As you can already see, this is a massive collaborative effort; fittingly so, given the massive scope of the problem, it’s trying to solve.
Building an open data ecosystem in the cloud
The core idea at the heart of this initiative is that we need to invert the traditional data sharing model, which has long relied on making datasets available on a download-only basis, meaning that researchers must download copies of the data to their institution’s data center in order to work with it. Now that the rapid technological progress in data generation techniques is fueling an extraordinary increase in the size of datasets being generated worldwide, reaching the order of petabytes per study, this model has become unsustainable: data transfer times become prohibitively long, and it is no longer reasonable to expect every research group or institution to maintain their own copies of these massive datasets.
The approach that has been proposed to solve this problem is to flip the script and, instead of bringing the data to the researchers, bring the researchers to the data. Specifically, by making the data available on cloud platforms, where it can be co-located with computing resources that researchers can use to analyze the data in place, without having to move any copies around.
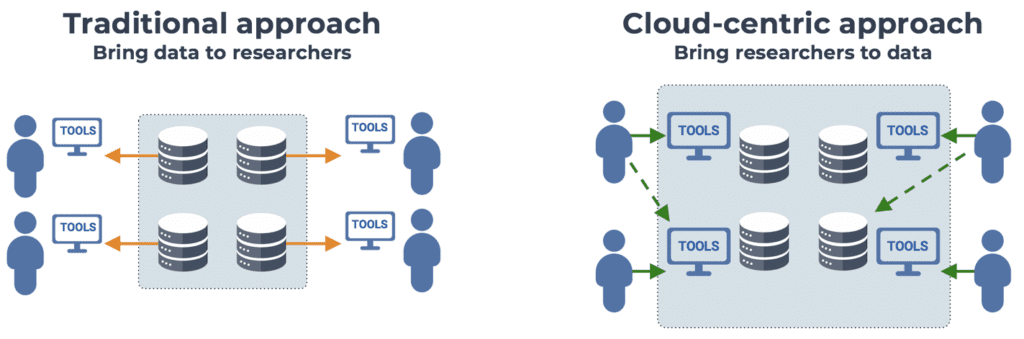

This cloud-based approach has many advantages, spanning topics from computational reproducibility to data security (hello FedRAMP), but perhaps the most exciting in my view is the prospect of making publicly-funded data more widely accessible to a greater number of researchers, particularly those who traditionally do not have access to massive data centers on-premises. Not only does this have great potential as an efficiency optimization hack (more bang for the federal data-generation buck) but it could increase fairness of the research ecosystem, which is an important goal in itself.
Yet the solution is not as simple as “just put the data in the cloud”. As fond as I am of the phrase “The cloud is just someone else’s computer that you rent,” the reality is that cloud infrastructure comes with its own specific complexities that prevent it from being readily usable out of the box by a majority of life sciences researchers.
So if we want the research community to be able to access and analyze data on the cloud effectively — without needing to get advanced engineering degrees on the side — it’s clear that we must also provide ready access to appropriate interfaces, applications, and whatever else is necessary to enable researchers to do their work as if they were using traditional on-premises computing resources.
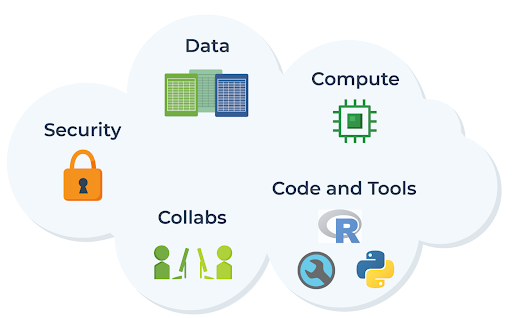

And that, in a nutshell, captures the spirit of what the AnVIL project aims to accomplish. As the full name “Analysis, Visualization, and Informatics Lab-Space” implies, the goal of AnVIL is to provide a virtual lab space that enables researchers to do all the computational work they need, end-to-end (spanning data processing, analysis, and visualization), in the cloud, with the same ease as if they were using traditional on-premises systems — with the added advantages of streamlined access to data, scalable compute resources, tools, and collaboration across organizations.
The AnVIL ecosystem viewed from Terra
To realize this vision of a cloud-based lab space for genomics, the AnVIL project pulls together five core components: Gen3, Dockstore, Bioconductor, Galaxy, and Terra. Within this context, Terra’s main role is to serve as a platform that brings together data access and analysis capabilities developed by the various partners involved in the project, as illustrated in the figure below.
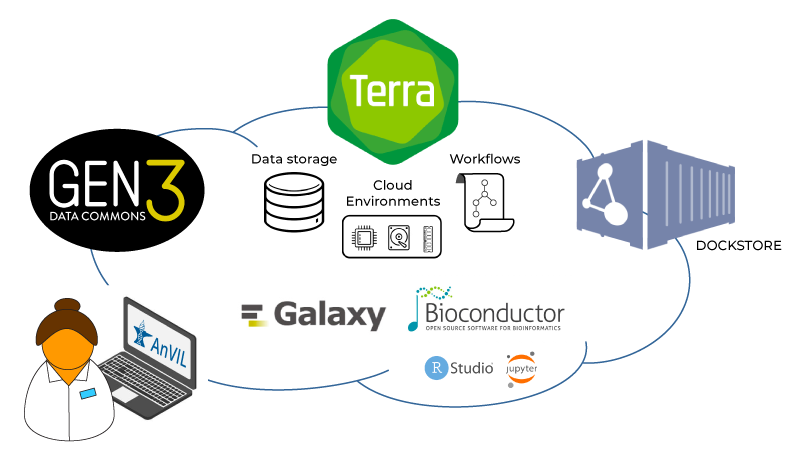

Key components of the AnVIL in relation to Terra: Gen3, Dockstore, Bioconductor, and Galaxy
Gen3 is a platform developed and operated by the University of Chicago that provides search and cohort-building functionality for AnVIL-sponsored datasets, which are stored in Terra workspaces, through a “data commons” framework. The Gen3 platform leverages GA4GH interoperability standards to provide export connectors to compatible platforms, including Terra. In plain English, this means you can browse the AnVIL data catalog, select data of interest, and export the relevant metadata to a Terra workspace. There, you can optionally combine it with other data (your own or other cloud-hosted datasets) and run analyses using the various apps and tools that are available in Terra.
For analyses that are composed of multiple steps and can be automated, Terra provides a scalable workflow execution system that uses Cromwell, a portable application developed by the Broad Institute, under the hood. The Cromwell-based system in Terra can tap into several sources of workflows including Dockstore, another AnVIL component, developed by the University of California Santa Cruz (UCSC). Dockstore acts as a community repository for portable analysis workflows that use containerized tools. Similarly to Gen3, Dockstore uses GA4GH interoperability standards to provide export connectors to compatible platforms, including Terra, so you can browse workflows in Dockstore and export them to a Terra workspace for execution.
For other types of analysis, such as those that require real-time interaction with the data, Terra provides a “cloud environments” framework that makes it possible to run certain popular applications within self-contained environments, such as Jupyter Notebooks, RStudio, and most recently, Galaxy.
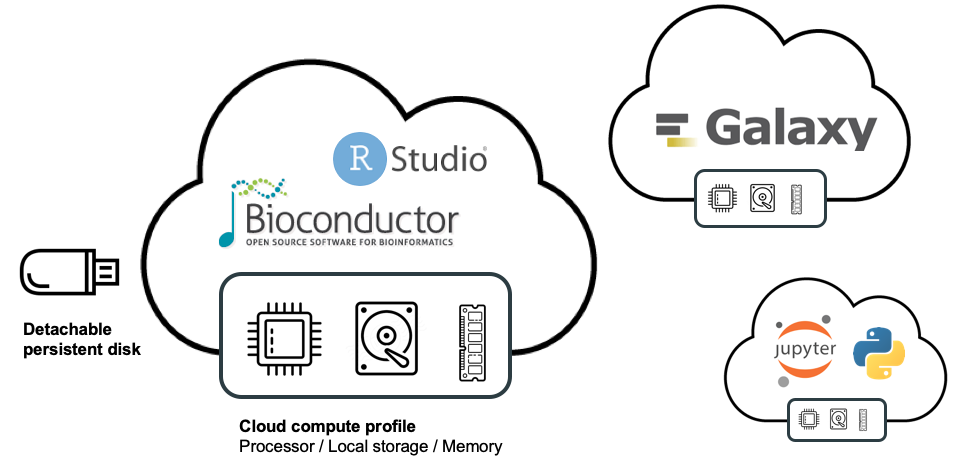

As part of the AnVIL project, members of the Bioconductor core team developed pre-configured Jupyter and RStudio environments that come preloaded with the core Bioconductor packages as well as an AnVIL-specific package, which we’ll cover in an upcoming blog post. This means you can launch either Jupyter or Rstudio in a few clicks and start working with Bioconductor right away because all the core packages are already installed. Similarly, members of the Galaxy core team developed a preconfigured environment that allows you to launch your own private instance of Galaxy with a few clicks, as we recently described in a previous blog post.
Note that all of these preconfigured environments are customizable, so you can install additional software on them as necessary; and you can also adjust the compute resources that are allocated to your environments if you need more processor power, memory or disk space. Overall the system is set up to provide out-of-the-box readiness for folks who are fine with the standard config while offering a ton of flexibility for those who have more specific requirements.
To learn more about how you can take advantage of all these resources, check out the “Getting Started” guide on the AnVIL project website, as well as the Events page which lists “popup workshops”, aka virtual tutorial sessions that can be very helpful for learning to use specific features.









