You may have heard that the cloud is the future of data sharing and collaboration. It’s all true! Moving data to the cloud is an essential step in enabling the research community to maximize the utility of reference datasets, biobanking efforts and other large biomedical datasets. By making data available on public cloud platforms, where they can be co-located with powerful and highly scalable computing resources, we can dramatically increase both the number of researchers who can access the data, and the ease with which they’re able to utilize it in their work.
Yet the core services offered by commercial cloud vendors are typically not usable out of the box by the majority of life sciences researchers, who don’t have advanced computing or engineering experience. In addition, sharing biomedical datasets requires specific access controls and querying functionalities that can be challenging to implement. To make this all work, it’s critical to provide appropriate tools and interfaces that “wrap” these core services and streamline data access and analysis operations for researchers.
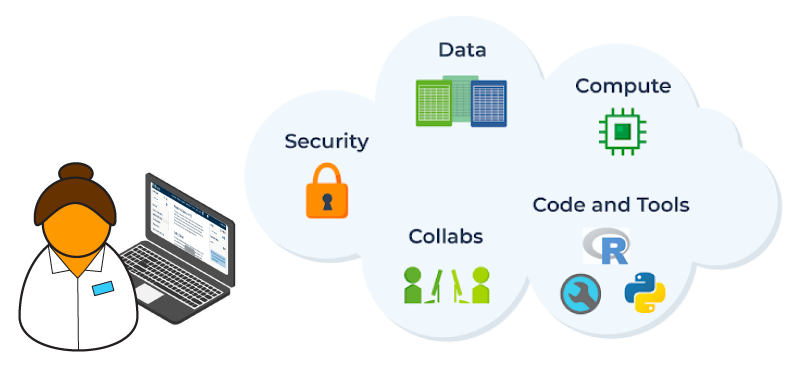

Cloud computing involves numerous, often complex components. Providing managed cloud services to researchers through interfaces tailored for their needs reduces burden and accelerates the discovery process.
That is why NIH agencies spanning multiple domains have commissioned infrastructure development projects that involve not just putting data on the cloud, but also building the additional layers of services that are necessary to deliver on the extraordinary promise of this new model for data sharing and analysis.
We’re proud that Terra plays a key role in several of these projects, including the NHGRI Analysis, Visualization and Informatics Lab-Space (AnVIL), the NCI Cancer Research Data Commons and its Cloud Resources (via Firecloud), and the NHLBI BioData Catalyst.
Each of these projects has its own specific goals, but they have a lot in common, both in terms of what they are building and how they are building it. They all involve multiple organizations collaborating to empower the community they serve, and follow some shared foundational principles.
Openness through interoperability standards
These projects all utilize technologies that are based on open standards developed by the Global Alliance for Genomics and Health (GA4GH), which means the resulting platforms and services, such as Terra, Dockstore and Gen3 for example, can be used in different combinations across these projects’ respective ecosystems. In a way, the projects are like specialized biomes that overlap in parts and together form a much larger, richer ecosystem.
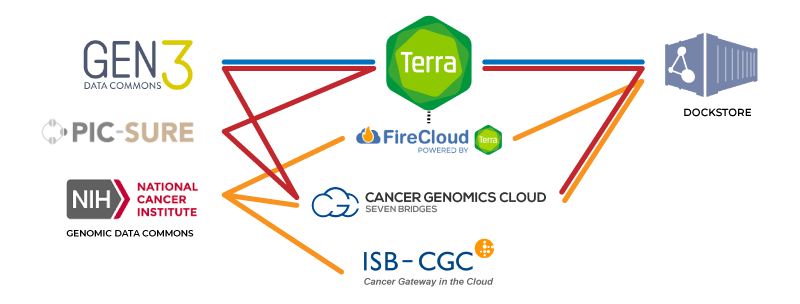

Simplified view of the connections between major components of the NHGRI AnVIL (blue), NCI Cancer Research Data Commons (orange) and NHLBI BioData Catalyst (red). FireCloud is powered by Terra, meaning all Terra functionalities are available in FireCloud.
As a result, researchers can often choose from multiple options what tooling and interfaces to use for analysis, depending on their preferences and their needs at the time, and they can later decide to switch to a different option with minimal friction and without losing work. This allows them to leverage the key strengths of each component platform or service without fear of being locked-in.
What’s more, because the underlying cloud platforms themselves are open, any organization is welcome to join the fun and add a new platform or service to the ecosystem; either to provide a tailored alternative to existing functionality, or to add novel capabilities.
Leveraging the insight of community experts
Speaking of capabilities, you might wonder how these projects decide what tools and services to make available to their respective communities. The answer is simple: by including people who are members of or even leaders in those communities. For example, the AnVIL project consortium includes the core development teams for Galaxy and Bioconductor. When we work with them to make those tools available in Terra, it’s a close partnership with the very people who have long been key contributors to those communities, and who know their needs better than anyone.
That leadership-based approach is complemented by various grassroots-focused initiatives designed to partner with individuals and groups in the relevant communities who are using the tools in their everyday work, and get their direct feedback about usability, pain points and unmet needs. This is a critical component of the infrastructure development process, and can take different forms across the projects. For example, the NHLBI BioData Catalyst project runs fellowship programs through which researchers can apply to receive funding in order to work with datasets such as TOPMed using one of the project’s analysis platforms. In exchange, fellows provide regular updates on their progress and any difficulties they have encountered, which is extremely helpful for the development teams to continuously adjust and improve the tools and interfaces.
Industry-grade security implementations and streamlined authentication
Finally, all these projects have in common a deep commitment to protecting the security of the datasets that they make available for research purposes, both as a philosophical stance, and as a requirement of the federal grants that support them. All the project components that handle access-controlled data are subject to stringent security audits in order to be authorized to operate in compliance with the U.S. Federal Information Security Modernization Act (FISMA). In the case of Terra specifically, we are proud to have recently achieved an even more stringent level of accreditation through the U.S. Federal Risk and Authorization Management Program (FedRAMP), which is shared by very few in our industry and widely acknowledged as a marker of excellence in information security.


Terra entry in the FedRAMP Marketplace. PaaS stands for Platform-as-a-Service. Moderate refers to the impact level of the Authorization to Operate that we received from FedRAMP, which is based on the type of data handled in the platform and the degree of severity that a data breach could entail.
There is of course a flipside to the security coin — we still need credentialed researchers to be able to work with the protected data, preferably without having to jump through a whole lot of hoops. The good news is that these projects use standardized authentication systems that are integrated with NIH systems, which streamlines access to controlled data comsiderably. Researchers simply link their NIH-issued credentials (such as ERA Commons ID) with their account on the project’s data platform; once this link is established, the platform mediates access authorizations behind the scenes, freeing researchers’ attention to focus on carrying out impactful analytical work.
And that freedom to focus, at the end of the day, is what these projects are all about.
From the vantage point of Terra development, our position at the heart of these overlapping biomes lends us an invaluable perspective into the needs of researchers across a wide spectrum of the life sciences. We are excited to continue building on the experience we have attained so far to support the growing number of communities that are joining the cloud data ecosystem through these and other initiatives.
To get started with any of the projects highlighted here using Terra as your home base, simply sign up for an account on app.terra.bio and follow the Getting Started instructions. For project-specific instructions and documentation, please see their respective homepages as linked above.



