Have you ever had to decipher a workflow written by someone else, and wished there was a quick way to figure out what it does and how? Earlier today, I gave a talk about this very topic at the 2022 Bioinformatics Open Source Conference (as part of ISMB 2022); and in the spirit of open-source, I’ve posted the pre-recorded version on YouTube!
In my talk, I presented a practical methodology for elucidating the structure and function of a workflow written in the Workflow Description Language, aka WDL. This systematic approach is intended to help bioinformaticians efficiently interpret and if necessary, reverse engineer existing WDL workflows. But first, a bit of context if you’re not already familiar with WDL.
What is WDL?
The Workflow Description Language (WDL, pronounced “widdle”) is a domain-specific language for describing data processing and analysis workflows. As such, it is not a full programming language; it is designed primarily as a means to define analysis tasks, chain them together in workflows, and parallelize their execution wherever possible. The language’s syntax strives to achieve portability across execution platforms, and lends itself especially well to execution on cloud infrastructure due to advanced support for containerization of individual steps. In addition, WDL is explicitly intended to be accessible to a wide range of users without requiring advanced programming experience.
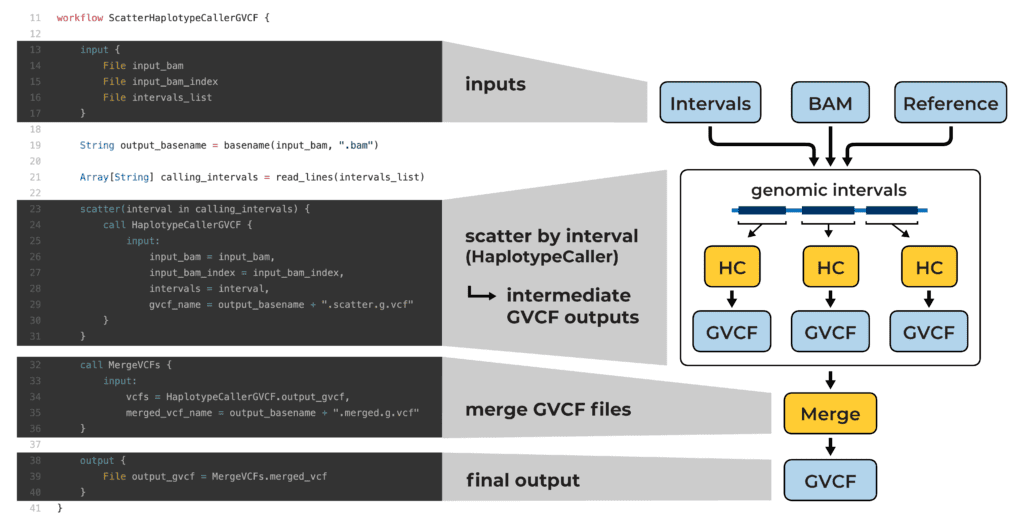

Example WDL script that parallelizes variant calling across genomic intervals (see blog post for details)
Originally created at the Broad Institute for developing GATK Best Practices workflows, the WDL language is now stewarded by the OpenWDL community, which was formed by individuals and organizations who adopted WDL in their own work.
The main sources of documentation and educational resources for learning WDL are provided by the OpenWDL organization on Github (BSD-3). Most notably, the WDL specification describes the syntax and features of the language (variable types, built-in functions etc.), and the learn-wdl repository provides introductory materials (including videos) explaining basic syntax and demonstrating execution through hello-world level examples and some sample workflows.
Reusable WDLs in the wild
Multiple consortia within the field of human genomics publish standardized workflows written in WDL, such as the Human Cell Atlas, and its adoption is spreading to non-human organisms and to related fields of study. For example, due to recent events, a growing number of public health laboratories in the US are adopting standardized WDL workflows for COVID-19 viral genomics and epidemiology.
Therefore it is increasingly likely for bioinformaticians to encounter workflows of interest written in WDL. Even if WDL is not their preferred language for developing their own workflows, bioinformaticians may need to be able to understand and apply these standard WDL workflows appropriately to their own data for continuity within a collaborative project, or they may want to reverse engineer and reimplement them in their preferred language.
A systematic top-down approach to deciphering a random WDL
Real-world, full-scale workflows such as the GATK Best Practices typically involve more complex patterns than what is shown in the OpenWDL hello-world tutorials. Deciphering such workflows is a very different exercise compared to writing one’s own workflow from scratch, and can benefit from employing a systematic approach, rather than attempting to read through the code linearly.
One popular form for documenting realistic and/or advanced use of a programming language is the “cookbook” style, in which the author posits a series of usage scenarios and shows how each would be implemented. This form is particularly helpful for developers who are interested in writing their own code. However, it is less helpful for someone who is trying to interpret an existing piece of code for reasons such as those outlined above.
In my talk, I presented an alternate approach to satisfying the latter use case. First, I pretend to stumble across an existing real-world workflow that is written in WDL but is not sufficiently documented, then I proceed to deconstruct it systematically in order to understand what the workflow is meant to achieve and how it does it.
The main take-home message here is the methodology itself, which can be adapted to other scenarios. As a secondary benefit, you will also gain some familiarity with WDL syntax and with interesting functional patterns of the language.
If you find this method useful and would like to see more WDL-related resources like this, please fill out the Terra support team’s WDL survey and let us know what you would find most helpful!








