Congratulations, your manuscript has been accepted for publication! Or it will be eventually, once you satisfy Reviewer #2’s latest nitpicks. In any case, you’ve spent months, possibly years pouring your heart and soul into this project, and you’re finally so close to the finish line. But… is finish really the right way to think about this? Hopefully, once your paper is out, others in your field will read it, find it compelling, and want to build on it — maybe even cite it! The question is, are you giving your readers everything they need to do so? For example, would they be able to reproduce key parts of your analysis, then apply the method to new data?
In last week’s Terra blog, I summarized a group discussion on the computational reproducibility of published papers that I led at the Bioinformatics Open Source Conference (BOSC) in July 2021, with the goal of identifying key challenges and solutions. Coincidentally, just this week Benjamin Heil and colleagues published a Comment in Nature Methods proposing a tiered system of reproducibility standards for publication of machine learning methods in the life sciences. Their recommendations, which include the use of containers and automated workflows as key instruments of computational reproducibility, are excellent and broadly applicable, not just for machine learning methods but for the publication of computational analyses in general. Some questions remain, however, when we consider how to enable the average researcher to implement these recommendations, which can be technically challenging in and of themselves.
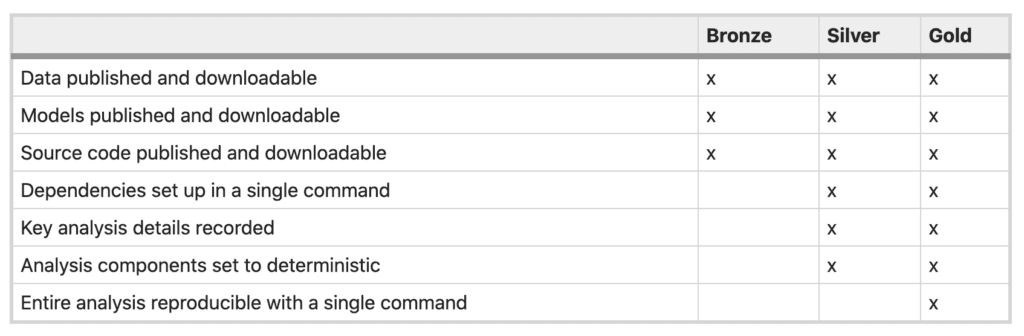

Table 1 from Reproducibility standards for machine learning in the life sciences (Heil et al., 2021)
One of the major obstacles we identified in our group discussion at BOSC was that, even given a full “list of ingredients” (data, tools, versions, command lines etc), it’s not necessarily trivial to pull them all together into a working environment that has everything set up the right way. You could satisfy the “gold standard” described by Heil et al. by putting workflow code in Github, one or more tool containers in Dockerhub, and data in a public bucket, yet your typical reader would still have a fair amount of work to do to be able to reproduce your analysis.
This is where Terra workspaces come in. As I’ll show below, the Terra workspace construct provides a really neat way to package your work and share it with others as a sort of interactive methods supplement that has the potential to significantly increase the computational reproducibility of your publications.
The Terra workspace as ultimate methods supplement
In a nutshell, a Terra workspace is a kind of computational sandbox that allows you to bundle together the data, code, and many elements of the computing configuration you use for your analysis into a single package that you can share securely with collaborators or publicly with the world.
The workspace also includes all the relevant logs, intermediate outputs, and so on produced in the process of running your analysis. Anyone you share it with can go in and examine how the analysis was set up, what the input files looked like, what all the parameters were, how long it took to run and what the outputs looked like, in full technicolor detail. So when you share a workspace, you’re sharing a lot more than just the ingredients and recipe for the cake; you’re also sharing a 360 view of the kitchen and much deeper visibility into the baking process.
You can go even further, and either give your collaborators access to use your workspace or just allow them to clone the workspace under their own account, which gives them access to a copy of the kitchen and the ability to do a full replay — on their own dime.

This is what gives the Terra workspace its exciting potential as an “ultimate methods supplement”. As you are preparing your manuscript, you can create a companion workspace that recapitulates your analysis, with all relevant metadata in data tables, and code set up to run either in Jupyter notebooks and/or as pre-configured workflows. If your dataset is access-controlled, you can set up your analyses to run on a set of public example data, with instructions for substituting a “real” dataset. Once you’ve run through the analysis at least once in this clean workspace, you can request* to have it made publicly readable. You can then include a link to the workspace in your manuscript, and once the paper is published, anyone will be able to view and clone your workspace. Readers will be able to examine and reproduce your work, and may even choose to adapt it for their own use.
* It is not yet possible for individual users to make their own workspaces publicly readable. To make a request, please see this documentation article.
Preprints, tool tutorials, and beyond
Then again, why wait for the formal publication when you could already associate a workspace with a manuscript preprint? That would enable other researchers in your field to examine your work and give you more informed feedback on your analysis before you even submit it to a journal, which would further the goal of using preprint servers for improving work shared prior to publication. Similarly, when you do submit a manuscript to a journal, you could provide reviewers with a fully loaded workspace, giving them the ability to evaluate your work more transparently.
More generally, this approach doesn’t have to be limited to publishing papers; it has a lot of potential for reducing the amount of human effort and time spent on “just getting things to work”. Several groups of computational tool developers are already using Terra workspaces to distribute their tools bundled with instructions, data, and configuration details in a way that makes them more widely accessible than just posting code and executables in Github.
As an example, one participant in our group discussion at BOSC recalled the evolution of the Broad Institue’s Genome Analysis Toolkit (GATK): from a standalone software package that you had to download, to containerized tools and pipeline scripts provided via Github and Dockerhub, and now full Terra workspaces where the pipelines are imported via Dockstore and staged with full configurations to run on example datasets, including full-scale whole genome data. (But yes, you can still download the GATK package if that’s what you really want.)
It’s worth reiterating at this point that Terra is not an isolated platform: it’s part of a whole ecosystem of open, interoperable platforms that are being developed to support the wide variety of computational needs that are emerging in the life sciences research community. For example, Terra and several other analysis platforms all connect to Dockstore, a standalone workflow repository that itself connects to Github. This allows tool developers to keep working in their own Github repositories, yet have their workflows be searchable centrally, and be available for seamless export to multiple analysis platforms, with a minimal amount of effort. So Terra workspaces are not “yet another silo”; on the contrary, they make it easier to bring together assets like containers and workflows that are traditionally deposited in separate repositories.
See it in action in the Terra Showcase
If this concept has piqued your interest, have a look at the Terra Showcase, which lists several dozen public workspaces spanning multiple types, from tool tutorials to method supplements. Many of these workspaces were developed primarily by tool developers and independent researchers who wanted to distribute their methods and enhance the computational reproducibility of their published work. (We recently redesigned the showcase to make it easier to browse and find workspaces based on your interests, so be sure to check it out if you had previously used the old version.)
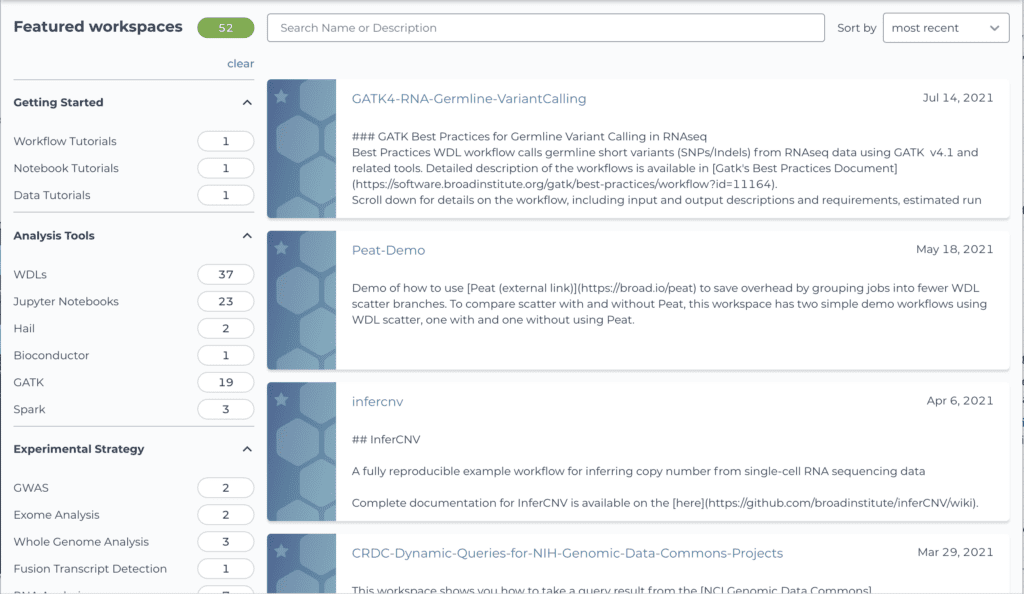

Screenshot of the recently redesigned Terra Showcase
If you like what you see, you can choose to clone the workspace under your own account and try running everything yourself — though you will have to pay the cloud vendor for the use of computing resources and storage for any files you generate or upload. The upside is that once you’ve verified the workspace works on the example data, you can pull in your own data to analyze, and/or modify the analysis parameters to suit your own purposes. This means you can go from reading about a cool analysis to trying it out for your own project in a record amount of time since you don’t have to install anything yourself. Then you discover something neat, publish your own paper with a companion workspace, and the virtuous cycle continues.
Resources
Heil, B.J., Hoffman, M.M., Markowetz, F. et al. Reproducibility standards for machine learning in the life sciences. Nat Methods (2021). https://doi.org/10.1038/s41592-021-01256-7
Working with Terra workspaces (Terra documentation)



