The 2021 Bioinformatics Open Source Conference (BOSC) held virtually a few weeks ago delivered a number of fantastic presentations, which you can now enjoy in their fully recorded glory on YouTube. Not included in those recordings, however, are the “Birds of a Feather” sessions, or “BoFs”, which are typically small, informal gatherings where participants have the opportunity to discuss topics of interest in a way that complements the main conference track. In this blog, I thought I’d share highlights and key takeaways from the BoF we organized on the theme of computational reproducibility of published papers, which drew a great group of BOSC participants and led to some lively discussion, in the hope that some of the ideas we exchanged would live on beyond the close of the conference.
“Think about the last time you wanted to reuse or adapt an analysis method that you read about in a published paper. How did it go? What were the biggest hurdles you ran into? What solutions are you aware of that you think everyone should know about, and what do you wish was available? Let’s talk about problems and solutions, and how we might make progress toward fully executable papers.”
— Conference program blurb
We all agree that we should build on prior work and avoid reinventing the wheel wherever possible. Ideally, we’d also like to avoid having to spend a ton of time figuring out how a published paper’s authors actually implemented the computational analyses they describe.
Yet we’ve all been stymied by computational methods sections that are just not complete enough to make it possible to redo the work step by step. For compute-intensive analyses, hardware requirements are often undocumented. Software version numbers and exact command lines are left out. Or, the command line is in there but some optional parameters were omitted despite making an important difference to the analysis. And of course, there’s the dreaded “the analysis was implemented with custom scripts (available on request)”.
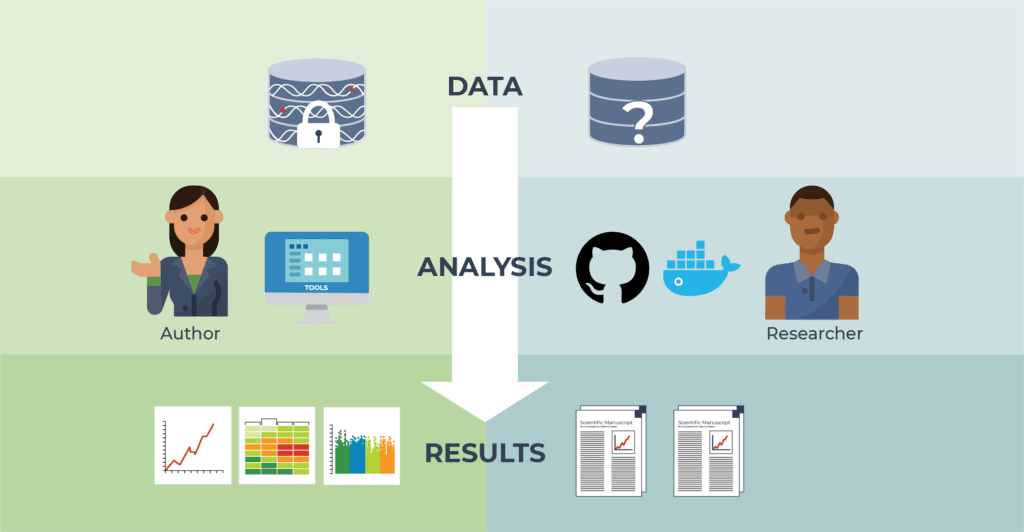

The reader of a published paper (pictured right) typically has an incomplete view of the full body of data, analysis method details, infrastructure configuration, and detailed results used by the author (pictured left).
Even granting that a lot of people are starting to pick up “good enough practices” like sharing code in Github, there is still typically a big gap between “I can download the code” and “I can get the code to run the same way you ran it in your study”.
It’s 2021. Why are we still dealing with these problems, and how do we get past them?
Incentives, or lack thereof
The first challenge that our group discussion converged on right away was that it can take a lot of effort to help others reproduce your own work, and that there’s a systemic lack of incentives for academic researchers to put in that effort. Even with the best intentions, many (particularly early-career) researchers are under intense pressure to get more work done —more papers published— so it’s difficult for them to prioritize an activity that is not explicitly rewarded in terms of career advancement.
Digging further into this, we found it useful to distinguish between two types of activities: there’s sharing a tool you developed to perform a particular type of data processing, then there’s documenting how you applied one or more tools in a study to solve a biological question. To be clear, it’s not just a matter of “tool developers vs. end-users” personas, because there’s a lot of overlap there — there are plenty of people who develop computational tools and apply them in their own research — and besides, many so-called “end users” typically generate their own code in the form of scripts that glue together invocations of command-line tools developed by others. That code in itself is an important, if sometimes undervalued and insufficiently shared, piece of the methodological puzzle.
Overall our little group expressed quite a bit of optimism about how things are going on the side of tool sharing activities; there are certainly plenty of challenges there, but also some great success stories. For example, the Satija Lab was cited as a model for their level of investment in documentation and support to empower the single-cell analysis community to use their Seurat toolkit. Based on some of our own experiences, we recognized that at least sharing the tools we develop can be associated with incentives rewarding community enablement, since for some types of grants, funders do attach importance to community uptake metrics such as the number of times your software has been cited or downloaded.
The situation felt bleaker at the other end of the spectrum, in regards to the computational reproducibility of methods in published studies. The only metric that seems to be clearly prized on the “output” side of scientific publishing is how many times your work has been cited, not whether anyone can actually reproduce the computational analysis you described (which is not the same thing as replicating/confirming findings). Intuitively we supposed that papers that present analyses that are more easily reproducible should get cited more often, by other investigators who were subsequently able to build on the work, but could not find any obvious support for this in the literature. It would be really interesting to see this be studied in a systematic way if only to be able to motivate paper authors to put in the work as a long-term investment.
We also discussed whether there is anything we could do as a community to reward investigators who do a great job of making their work more readily reproducible (especially early-career folks). Perhaps through a new award by the Open Bioinformatics Foundation, if sponsors could be found to support such an initiative?
The non-linear nature of the research process
Moving on from getting mildly depressed about how academic incentive structures are all wrong, we reflected on another challenge: the conflict between the reality of how most scientific investigation proceeds — on a winding path, with many branches and failures along the way — and the necessary exercise of rewriting history in order for the paper to present a coherent narrative.
We discussed the idea of doing more to show the messiness and the failed attempts that hide behind what is effectively a redacted sequence of events presented in the eventual paper. Greater transparency would be especially helpful for training students and newcomers to the field, and giving them a more realistic understanding of “how it all really works”. One participant mentioned that their lab shares unredacted notebooks in Github that track everything that happens in every project, for full transparency.
Ultimately though, for the purpose of empowering others to build efficiently on our published work, we agreed it does make sense to compile a “cleaned up” version that explains linearly what to do to reproduce the work — in the narrow sense of recreating the same outputs based on the same inputs, with the same tools.
What generally stands for this in an average paper’s methods section is a text description of what was done, perhaps with some supplemental materials providing additional details about the software, links to code in Github, and specific command lines if we’re really lucky. Yet as we discussed earlier, that is almost always insufficient, because key details tend to get lost in the process of rewriting the history of the analysis. This is especially the case when multiple contributors were involved, and the person in charge of compiling the materials may not themselves fully understand all the intricacies of every step in the computational journey that they are reporting.
Here’s a provocative idea: perhaps every author (first? corresponding?) should go through the exercise of redoing the computational analysis their paper describes based only on their own methods section. That should encourage them to provide step-by-step commands and executable assets rather than prose alone, and would go a long way toward ensuring that their methods section is in fact sufficient to reproduce the work in a way that is equivalent to how it was originally done. (Perhaps on a downsampled or truncated dataset if cost or runtime were to be a major obstacle.)
So, daydreams aside, assuming we could motivate people to do more to make their computational work reproducible, what are the technical opportunities that could grease the wheels from a practical standpoint?
Computational sandboxes in the cloud
We haven’t talked about actual computing infrastructure until now because a lot of the problems we face here are fundamentally human problems — misaligned incentives and gaps in accountability. Yet infrastructure has to be part of the solution because it is the remaining piece of the problem: we often struggle to reproduce computational work because of the heterogeneity of the computing environments that are available to us, and the complexity involved in installing and managing software packages.
This suggests an important role for cloud-based infrastructure, which enables us to share software tools, code, and data in pre-configured environments where everything just works out of the box.

The group brought up several cloud-based platforms like Code Ocean, Gigantum, Binder, and Google Colab, which are all popular solutions for sharing code in cloud environments that provide at least basic computing capabilities for free (typically with paid tiers that offer better performance). This allows you to do things like test-drive software tools and code solutions without installing anything yourself, and share your own tools and code with others.
In fact, some journals are now working with Code Ocean and others to enable reviewers to actually run code described in manuscripts without having to do any installation legwork. This presents the exciting possibility that evolving peer-review standards — such as requiring code to be made available on such a platform as part of the manuscript submission— could lead to a higher quality of published software tools.
One limitation of the free version of the resources provided by these platforms is that they are backed by fairly minimal virtual machine configurations, which typically can’t handle the scale of the new generation of big-data analysis domains like genomics. They’re great for teaching and for toy examples, but they’re not what we need for full-scale work.
The other important limitation is that their features tend to be rather generic; they’re not designed to support bioinformatics work, nor to accommodate the needs of an audience of researchers who may have an advanced understanding of complex statistical analyses, but minimal training in using cloud computing infrastructure.
So what other options are there? We wrapped up the discussion by talking about initiatives like AnVIL that aim specifically to empower the life sciences research community to take full advantage of the power of the cloud, in combination with access to relevant datasets and tools through interfaces tailored to their audience. By supporting the development of platforms like Terra, Dockstore, Gen3, and others, infrastructure projects like AnVIL solve the problem of the heterogeneity of what hardware and software environments people might have access to and provides a venue for staging full-scale analyses in a way that can be shareable and work out of the box for anyone.
Yet there is a lot more to say about how Terra as a computational platform offers enormous potential for supporting and promoting computational reproducibility, particularly for published papers. In the next Terra blog, I’ll lay out some relevant ideas as well as concrete examples of efforts that are already underway to use public workspaces as “the ultimate methods supplement” to make papers effectively executable, and more besides.








