Hot on the heels of R Shiny support, our Interactive Analysis team has just released an upgrade that enables adding Graphical Processor Units (GPUs) to your Notebook cloud environments in Terra.
GPUs were originally developed for graphical processing, as the name implies, and rose to prominence as a way to accelerate 3D computer graphics for applications like video games, which involve a ton of math that has to be done very quickly. Specifically, linear algebra operations like matrix multiplications, which happen to be the same kind of computations involved in a lot of machine learning (ML) algorithms.
Fast forward a decade or two, and there is now a vibrant ecosystem of ML tools and packages that make use of GPUs to accelerate computations in a wide variety of domains, including the life sciences.
How to use GPUs in Terra
Terra has offered the ability to use GPUs in workflows for quite some time, yet many of you have told us that you also need to be able to run GPU-enabled computations interactively, so we’re really excited to roll out GPU support for Jupyter Notebooks.
To create a Notebook cloud environment with GPU(s) attached, simply check the “Enable GPUs” option in the environment configuration panel. This will bring up two additional menus from which you can select a specific type of GPU, as well as the number of GPUs you want, as shown in the screenshot below.
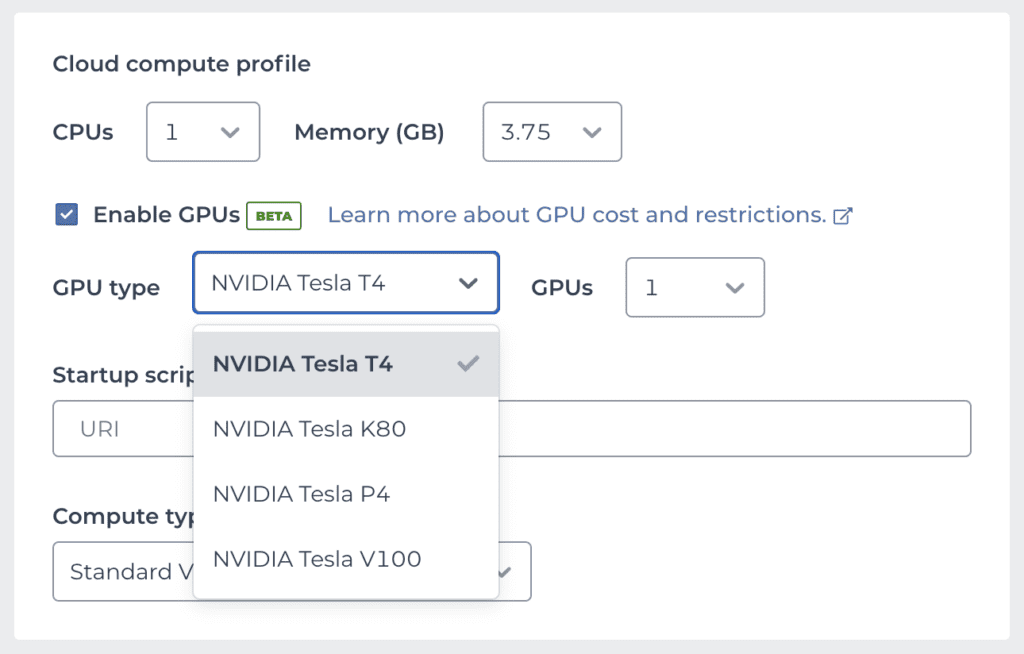

The Tesla T4 is currently the most popular model of NVIDIA GPUs on Google Cloud for ML applications. As a result, T4 instances may become unavailable during peak utilization periods. If your environment creation request fails with an error about resources being unavailable, see the documentation for workarounds.
Note that if you already have an existing cloud environment configuration, you will have to delete it explicitly before you can set up the GPU-enabled configuration, as documented here. This is a bit different than how the cloud environment configuration options normally work, but we expect this to be temporary — the GPU support feature is currently in beta status, and the development team has plans to address this and some other minor limitations in upcoming work.
GPUs in action for single-cell analysis
You can try out this new functionality with a Python analysis package called Pegasus, a component of the Cumulus framework developed by the Li lab to analyze the transcriptomes of millions of single cells. Pegasus features several algorithms that can optionally take advantage of GPUs: a Pytorch-based implementation of Harmony (Korsunsky et al., 2019), which applies a batch correction in order to integrate single-cell data from a variety of experimental sources; and two types of non-negative matrix factorizations using the nmf-torch package.
Conveniently, Bo Li and colleagues maintain a public Terra workspace named Cumulus that contains several Jupyter Notebooks as well as a cloud environment image that contains all the software dependencies required to run Pegasus, which is available as one of the preset community-maintained environments. This means you can take Pegasus — and a GPU! — for a spin with just a few clicks: clone the Cumulus workspace, then set up your cloud environment by selecting the Pegasus image and enabling GPU support as outlined above. Your configuration panel should look something like this:
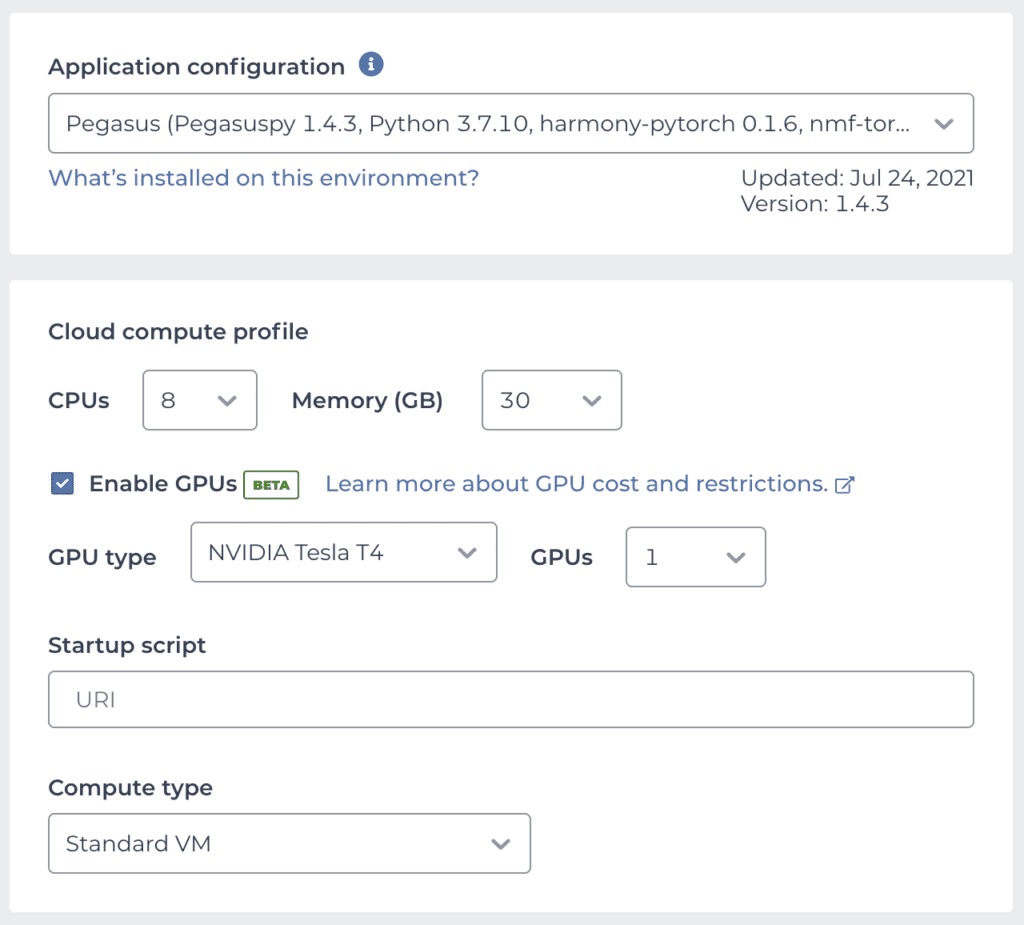

We’re using 8 CPUs with 30 GB of memory to provide a solid compute base for the other analysis steps that can’t use the CPU. For the GPU, we’re selecting the Tesla T4, which is the most widely recommended option for ML from NVIDIA’s lineup.
Once your environment is running, open the notebook named “GPU Benchmark” in the Notebooks tab and run all the cells. Scroll down to the Batch Correction section and you’ll see two cells that run two slightly different versions of the same “run_harmony” command, each on a copy of the same original dataset.
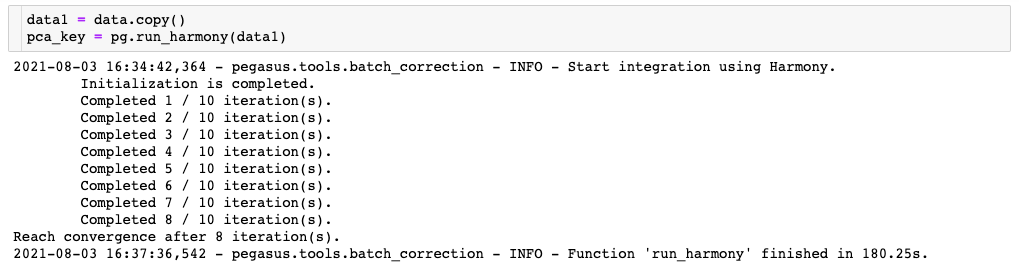
In the first one, the Harmony algorithm uses the CPUs only, even though a GPU is available in the environment because use of the GPU is not explicitly enabled in the command:

Compare this to the second one, where the Harmony algorithm is able to use the GPU thanks to the addition of the “use_gpu=True” parameter:
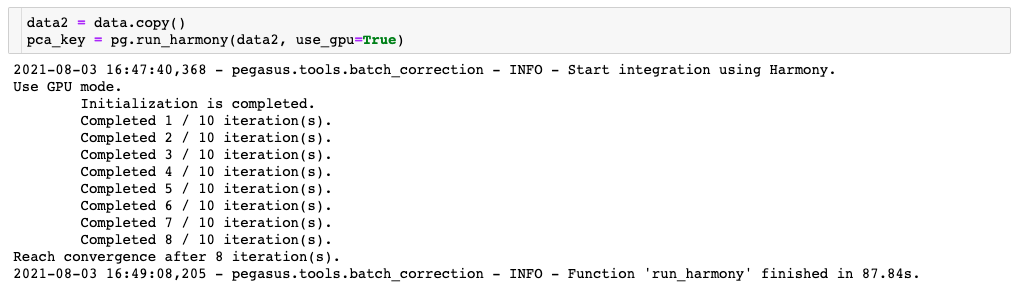

As you can see, the GPU-enabled version ran more than twice as fast as the CPU-only version of the command. The CPU-only configuration is quite a bit cheaper, but since it has to run for a longer amount of time, you may end up paying similar amounts regardless of what you choose. The tiebreaker to this tradeoff then, is this: what is the value of your time?
If you try to run the GPU-enabled version of the command on a CPU-only configuration, the Pegasus software will warn you that “CUDA is not available on your machine” and it will fall back to using the CPU-only mode. CUDA is the programming interface that software tools like Pegasus use under the hood to access GPU capabilities.
Finding the sweet spot of performance vs cost
For completeness, we ran this again several times to test the range of CPU-only configurations available in the system and see how performance scales with the number of CPUs we throw at the problem. We also tested the other available types of NVIDIA GPUs to see if everyone was right about the Tesla T4 being the best option for this kind of work.
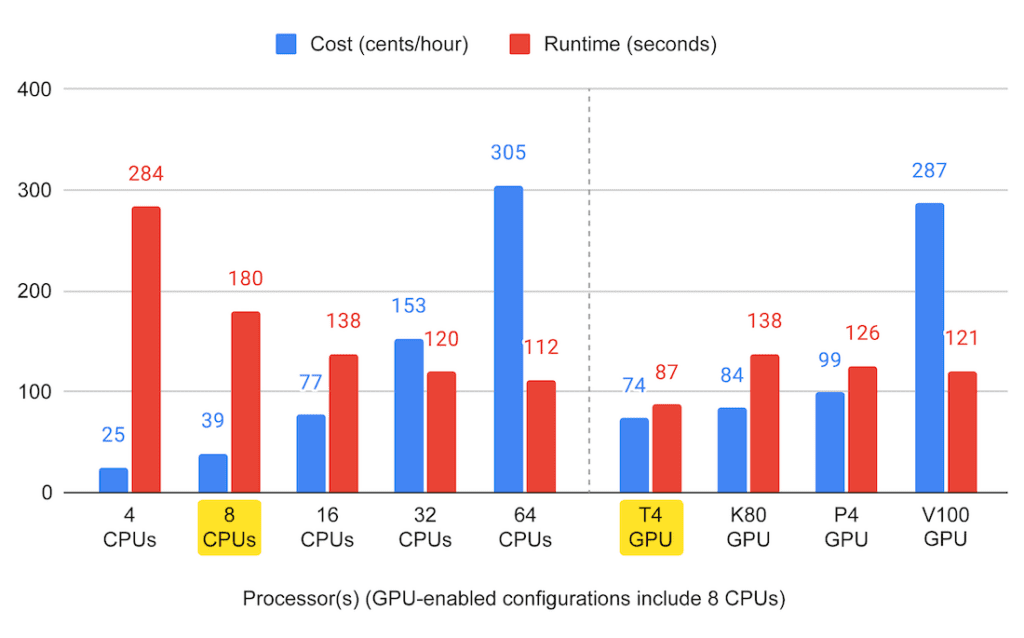

This summary graph suggests that you can certainly get pretty far just by adding more CPUs, but CPU-only performance quickly reaches a plateau (though cost does not, oh no — that just keeps going up). And yes, out of the available NVIDIA machines, the Tesla T4 does look like the right option for this job, with the lowest price point and lowest runtime, a rare combination indeed.
Keep in mind of course that this is a fairly informal benchmark of just one algorithm implementation, using a dataset that is not small but also not enormous. We did see some variability in the runtimes, which may be due to latency in the data handling and so on, so your mileage may vary.
More generally, what you see here does not demonstrate the full scale of the acceleration that you can reap from using the right GPUs in the right place. Consider it more of a teaser meant to motivate you to try out this new feature… and as always, let us know how it goes!
Resources
Korsunsky, I., Millard, N., Fan, J. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). https://doi.org/10.1038/s41592-019-0619-0
Li, B., Gould, J., Yang, Y. et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat Methods 17, 793–798 (2020). https://doi.org/10.1038/s41592-020-0905-x
Yang, Y. and Li. B. Harmony-PyTorch: a PyTorch implementation of the popular Harmony data integration algorithm. https://github.com/lilab-bcb/harmony-pytorch



