One of the most heartening current trends in the life sciences is the increasing abundance of consortium-based research projects that are explicitly committed to generating large data resources and sharing these with the wider research community. There’s a lot to say about how this approach can maximize impact, reduce duplication of effort and increase participation across the board — but what better way to make that case than by a concrete example.
Specifically, let’s take a look at the BRAIN Initiative Cell Census Network (BICCN), a research program within the NIH’s BRAIN Initiative that seeks to identify and catalog the diverse cell types in human, monkey and mouse brains. The program involves a consortium of research teams at over a dozen organizations who are conducting different projects looking at various aspects of brain development, with a variety of experimental strategies and assays.
The BICCN program recently reached its first major milestone: the completion of an effort to produce comprehensive mapping of mammalian primary motor cortical cell type identities on a molecular level. This milestone is marked by the publication of a “Flagship Release”, ie a main paper describing the overall project and findings, as well as a Nature “Collection” that indexes key studies published so far in the journal by consortium members, accompanied by an editorial and a handful of assorted news features. Some of the member organizations have also written up their own perspectives; for example this Broad Institute news article about the work of Evan Macosko and colleagues.
Save the date: There will be a live webinar on Wednesday, October 27, 2021 featuring lead investigators from the BICCN consortium who will discuss their findings.
What I find really exciting however is that this effort goes beyond “just” being a collection of papers from consortium members. The BICCN program dedicates significant effort to making the data and tools used in their papers widely available to the research community, so that others can reproduce and build upon the work presented in the papers.
For example, they have built a data repository called the Neuroscience Multi-Omic Archive, aka NeMO, that provides access to all the transcriptomic and epigenomic data generated by BICCN investigators. Importantly, this is not your typical data download portal — the data is hosted on Google Cloud, and although it is possible to download the files, the portal is designed to facilitate using the data on the cloud directly, without having to download and maintain your own copy.
From NeMO to Terra
The NeMO portal includes a data browser interface that allows you to explore and select BICCN data using various criteria — organism, assay technology, investigator and so on — with a “shopping cart” paradigm inspired from online stores. When you’re ready to “check out”, the portal provides options to either download a file manifest or sample metadata, or export the data to Terra for analysis on the cloud.
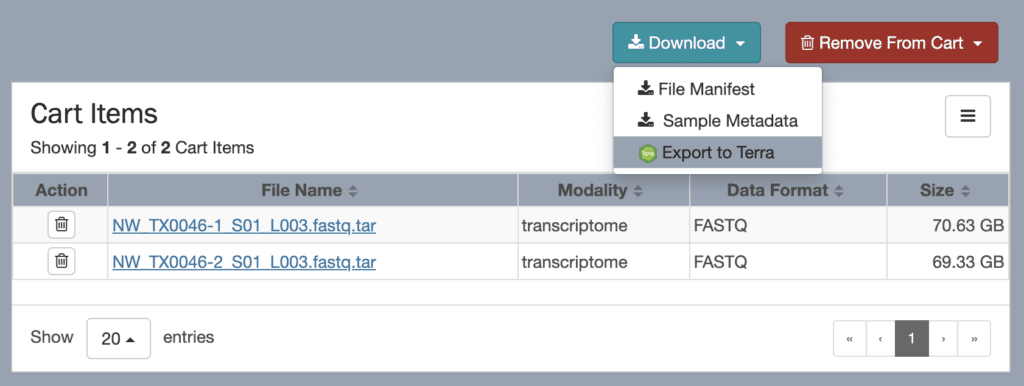

Crucially, this export operation does not create a copy of the data that you would then have to maintain and pay to store. Instead, it exports the metadata, which includes links to the original locations of the data files in the NeMO repositories, to a Terra workspace.
Once there, you can run your own analyses on the data, either through Terra’s highly scalable workflow execution system or through one of its interactive cloud environments. These come preloaded with applications like Jupyter Notebook and RStudio, as well as popular tool suites like Bioconductor, and can be further customized by installing additional packages. You can integrate the data with your own, or with data from other cloud-based projects and repositories that are connected to Terra in the same way.
Finally, you can publish results (or simply share them privately with collaborators) in the Broad Institute’s Single Cell Portal, which is connected to Terra and provides convenient data exploration and visualization options.
Get started today
To help researchers get started with BICCN data and tools, the NeMO and Terra teams collaborated to create a tutorial workspace in Terra that includes instructions for importing example transcriptome sequencing data from NeMO (in FASTQ format) and run a few preconfigured analyses: aligning the data, producing a raw count matrix with quality metrics; filtering, normalizing, and clustering the raw count matrix; and finally, exploring the resulting single-cell data with Seurat in a Jupyter Notebook.

The NeMO and Terra teams also deliver a variety of workshops online and at conferences, for which the materials are generally posted online. You can find the latest BICCN Omics Workshop materials on the NeMO website; make sure to click the “Tools Covered” tab to get a more detailed sense of the content, and the “Recordings” tab to access the videos. You’ll see the resources provided cover a number of topics and tools beyond the NeMO-Terra connection and analysis capabilities.
The video covering the Terra portion of the October 2021 edition of the workshop (aka “Day 2” in some descriptions) is available here. There is a version from January 2021 that is also applicable if you wanted to get started with that version. That session was given by the Terra team and is documented fully here. It introduces the Terra platform and demonstrates step-by-step how to work through the instructions provided in the tutorial workspace to analyze data from NeMO, and how to publish results in the Single Cell Portal.
In light of the BICCN’s recent Flagship Release, it’s tremendously exciting to see these extensive resources being made so readily accessible to the wider research community, and we look forward to seeing the many follow-on studies that will build on top of this work.



