I recently had the pleasure of giving a presentation at the Google Cloud Next ’21 conference, summarizing some of the work that the Broad Institute’s Data Sciences Platform group has done in collaboration with Intel and Google Cloud to scale up our genomic analysis capabilities and share the benefits with the wider research community. This blog is a lightly adapted version of my talk, which you can watch on YouTube if you prefer video.
Our mission at the Broad Institute is to uncover the molecular basis of major human diseases to inform prevention, detection, treatments and cures; develop effective new approaches to diagnostics and therapeutics; and disseminate discoveries, tools, methods, and data openly to the entire scientific community. In the course of fulfilling this mission, we face many challenges. One of them is the deluge of data that we’ve been experiencing in recent years.
Extraordinary improvements in data generation technologies and plummeting costs over the past decade have led to a boom in the number of datasets that are being produced. That gives us an unprecedented opportunity to fuel scientific breakthroughs and deliver on the promise of precision medicine.
Yet all that data is only useful if we can analyze it at scale, collaboratively.
What’s hard about that?
The most immediate problem we face is dealing with the scale of data processing involved. We’re generating terabytes of data every day, and every byte of data has to go through massive pipelines designed to squeeze out the useful information from the raw signal. The longer it takes for those pipelines to run, the more time we have to wait between generating the data and deriving actionable insights.
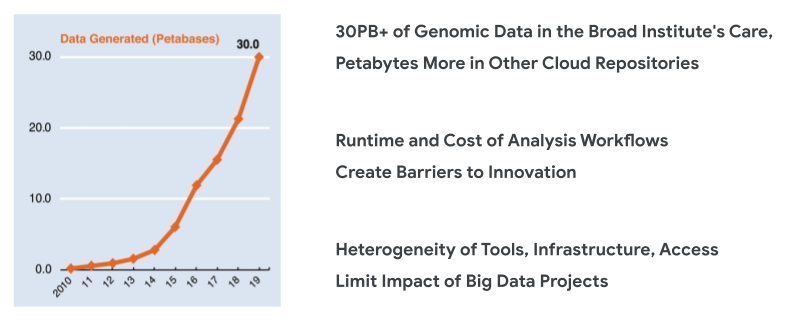

And then there’s the question of cost. Even when the cost of a single operation is small, these small costs per job can add up to huge sums of money when we’re looking at cohorts of hundreds of thousands of human genomes. Every extra cent that we spend on a pipeline that is not as efficient as it can be turns into a limitation on the scope of the research we can afford to undertake.
But that’s still not all. Frankly, there’s so much work to do to maximize the use of these datasets, and not enough brains in the game yet. We need to broaden the range of researchers who can effectively contribute to this work. It can’t be the province of only a handful of big established powerhouses. There are lots of scientists throughout the world who could contribute so much more if they had access to the right data and tools.
So how do we tackle these obstacles?
Solving genome-scale challenges collaboratively
For the past 5 years or so, we’ve partnered with Intel through the Intel-Broad Center for Genomic Data Engineering. It’s been a wonderful collaboration that has produced robust solutions for accelerating our tools and pipelines. These have helped us take control of the flood of data, scale up our analyses and, with some extra help from Google Cloud engineers, reduce our costs when we moved the institute’s genomic analysis operations to the cloud.
As a complement to that work, we’ve been co-building the Terra platform in partnership with Microsoft and Verily, which enables us to put the power that these optimized tools deliver into the hands of just about any researcher on the planet.
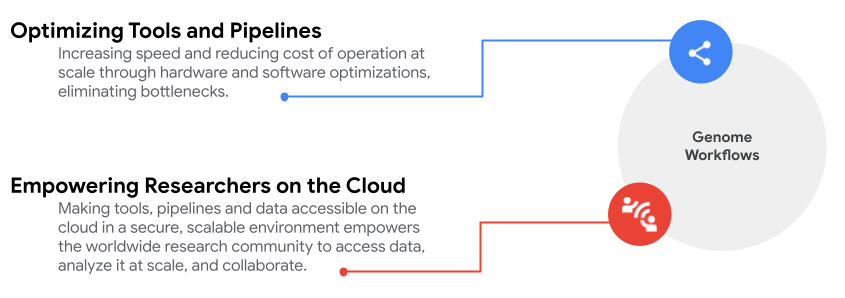

Read on for a bird’s eye view of these collaborative efforts.
Optimizing tools and pipelines
We use many different tools for genomic analysis, but none so much as the Genome Analysis Toolkit, or GATK, which we develop in-house and make available to the research community as an open-source package. It’s the world’s most widely used toolkit for identifying variants in short-read genome sequencing data.
We run it in the form of automated pipelines that will take a massive pile of raw sequence data and put it through several layers of transformation to extract the genomic variants. Those variants are what researchers ultimately care about for understanding human genetics, diagnosing diseases and eventually, designing strategies for treating or even eliminating those diseases.


But all that processing is a lot of work, and there are some classic bottlenecks that for a long time really restricted our ability to scale up our analyses.
Thankfully, our Intel collaborators developed optimized versions of some of the key algorithms involved, which now run faster on chips equipped with features like AVX-512. We also had the opportunity to collaborate with engineers at Google Cloud who helped us optimize our pipelines to take advantage of specific features of their cloud architecture, delivering a whopping 85% reduction in cost compared to our initial (naive) implementation.
Our optimized pipelines are fully open-source and available in the WARP repository on Github.
More recently, we’ve seen a general acceleration of processing with the latest generations of CPUs. For example we recently performed some benchmarking that showed faster execution times on the 2nd generation Xeon processors, again with some really significant effects on classic bottlenecks like genome alignment and variant calling.
When we ran the benchmarks on Google Cloud, comparing N1 instances equipped with older processors against N2 instances equipped with the Xeon family of CPUs, we saw the N2 instances outperforming the N1s on both runtime and cost — so the pipelines were faster and cheaper to run. Check out this previous blog post to learn more about the benchmarking protocol and our findings.
Empowering researchers on the cloud
I mentioned it’s imperative that we get more brains into the game, and the cloud is an obvious way to increase access to large datasets and put more computational power into the hands of researchers across the world. But we know from our own experience that you can’t just throw life scientists and physicians at the Google Cloud console. There’s too much “IT stuff” to navigate, and training researchers to deal with it directly would be far too onerous considering their time constraints and priorities.
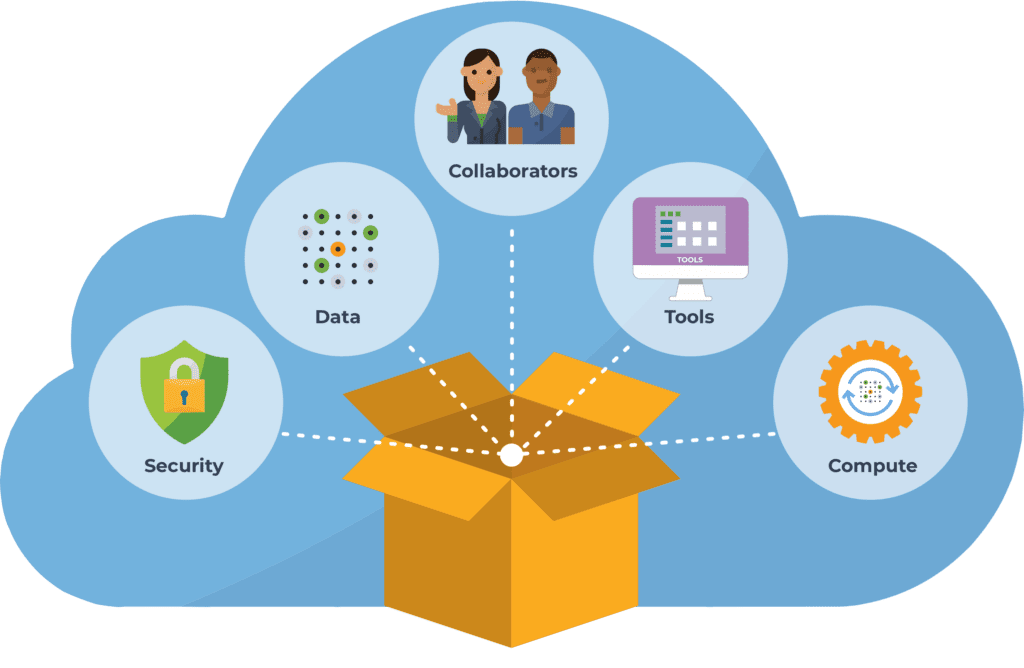

That’s why we partnered with Microsoft and Verily to develop Terra as a platform that sits on top of the cloud infrastructure, and packages the most relevant functionality in a form that is more immediately usable by researchers in the life sciences. Terra provides applications and interfaces that are tailored to the needs and expectations of life scientists; and it connects together the data, tools and computing resources needed to advance biomedical research.
Importantly, since we’re talking about human biomedical data, the entire platform operates according to stringent standards of security under the FedRAMP program, as we’ve discussed in this previous blog. So a credentialed researcher can easily access the datasets they need, and apply the tools they want, securely and at scale. And, they can share their work at any stage, either just with their collaborators or with the world, in a form that makes their analysis completely reproducible and extensible.
Yet, it’s not only about the researchers. Terra brings together a multitude of people and organizations with different roles to play. Funders, data generators — even patients who wish to donate their data to research — all want more researchers to be able to make use of their precious data.
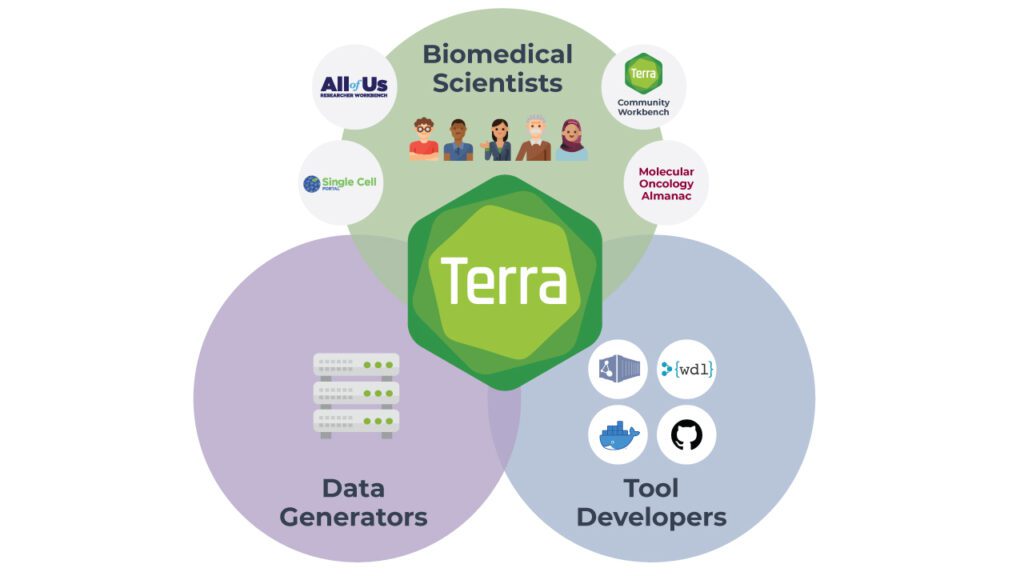

Meanwhile, tool developers want to make their tools available to the research community in a way that just works out of the box, with containerized tools, example data and preset configurations, and a minimal amount of support burden.
For example, the optimized GATK tools and pipelines we developed with Intel are available in Dockstore and pre-loaded in Terra for anyone to use. That means any researcher in the world can take advantage of the Intel team’s optimization work to run scalable analysis pipelines that are fast, cost-effective and scientifically excellent, without having to install anything themselves. This maximizes the impact of our collaborative problem-solving; and any other group can do the same with their own pipelines, tools or other resources they wish to share with the community.
A data ecosystem that fosters collaboration
Finally, I want to emphasize that Terra is not a walled garden. It’s a hub in an open ecosystem of platforms, portals and data repositories that are interconnected through open-source standards, following the vision originally laid out in the Data Biosphere manifesto. You can learn more about our collaborative approach and our many partners here.
If you’d like to join us in this brave new world of genomics in the cloud, please visit Terra.bio and take the GATK whole genome analysis pipeline for a spin. Or, browse the multitude of preloaded workspaces in the Terra showcase to find other use cases that might interest you.
See you on the cloud!



