Tim Looney and Vanesa Braunstein are co-authors of this blog. Tim Looney is the Senior Director of Scientific Affairs at Singular Genomics. He has a background in NGS assay development and bioinformatics. Vanessa Braunstein is the life science product marketing manager at NVIDIA focusing on genomics and drug discovery. She has a background in molecular and cell biology, genetics, pharma, and marketing.
DeepVariant is a CNN-based accurate variant caller for germline workflows of short and long-read data. It is part of NVIDIA® Parabricks®, a suite of GPU-accelerated and AI-based genomic analysis applications for high-throughput DNA and RNA data. Parabricks includes a variety of optimized and AI-based industry-standard genomic tools, which deliver up to 107x acceleration over CPU-based tools and reduce compute costs by up to 60%. To put these numbers into context, this means that a 30x whole genome can be analyzed in just 16 minutes compared to ~24 hours on CPU. That translates to the analysis of up to 30,000 whole genomes a year on a single server.
The newest version of Parabricks (v4.1) now offers a new DeepVariant retraining tool which enables researchers to fine-tune DeepVariant to their specific lab protocols, enhancing the accuracy of variant calling in genomic analysis workflows. This is important, as subtle and unique artifacts can be introduced to samples from a variety of sources, such as varying versions of wet lab kits, reagents, and sequencers used by different labs.
In this blog post, we will discuss how the DeepVariant retraining tool was used on Singular Genomics’ sequencing data to improve variant calling accuracy.
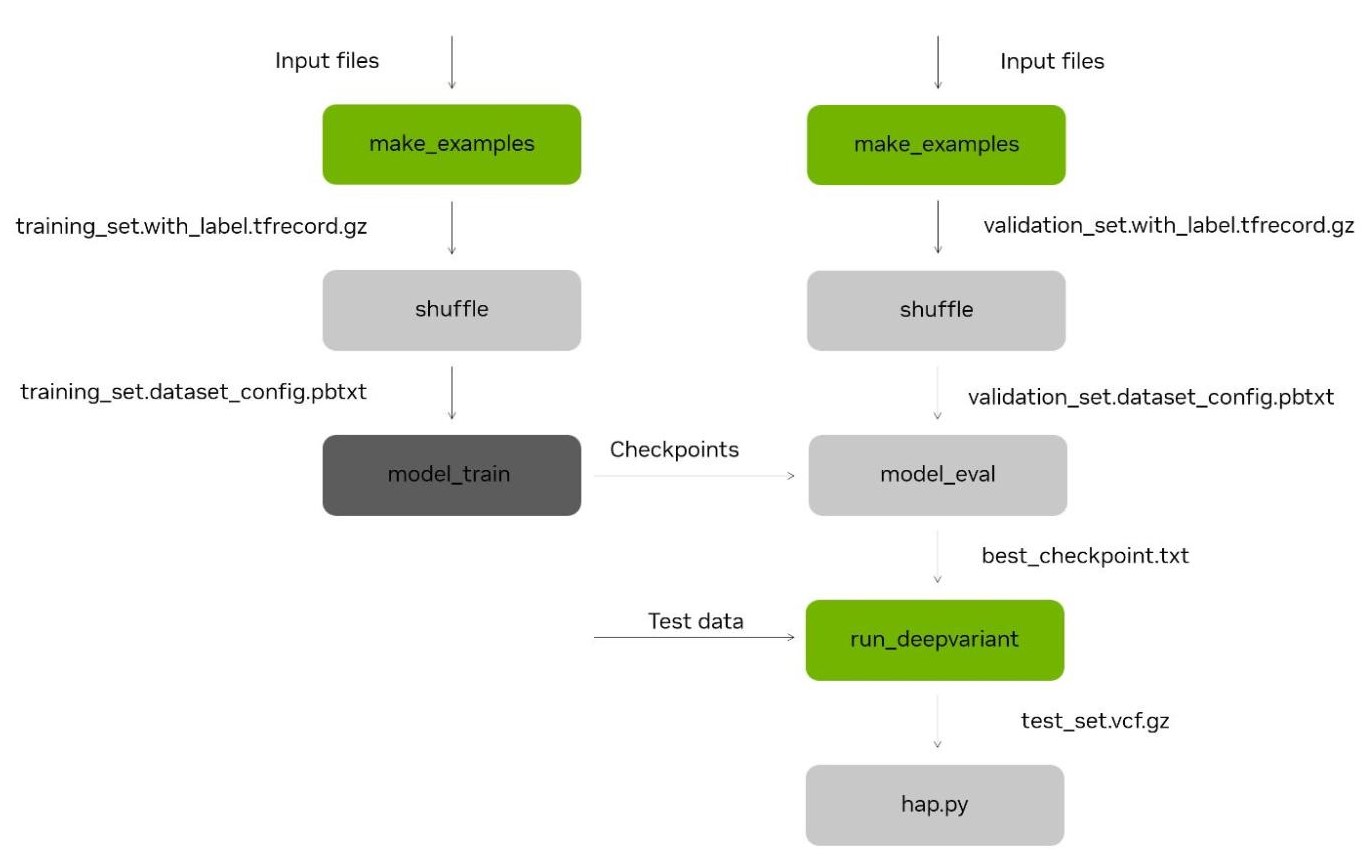

Figure 1. The DeepVariant re-training tool involves generating candidate variants in pile-up format (`make_examples`), before shuffling and using to train (left) and test (right) the DeepVariant CNN model. This model can then be used with DeepVariant, for variant calling inference (`run_deepvariant`) and accuracy testing with hap.py. The stages `make_examples` and `run_deepvariant` (green) have been accelerated as part of the NVIDIA Parabricks software suite.
Singular Genomics has been working to build a next generation sequencing device capable of delivering faster, more flexible sequencing. In 2022, the company launched the G4 sequencing platform, which can deliver up to 1.6 billion 2x150bp reads in under 24 hours, which makes it one of the most powerful benchtop systems. To achieve this milestone, a rapid sequencing-by-synthesis chemistry was developed, including novel sequencing enzymes and optimized fluidics, along with a powerful image processing solution to keep up with G4’s fast sequencing. To power this, Singular utilized NVIDIA GPUs for AI-driven base calling due to the flexibility and speed offered by the GPu architecture.
Given Singular’s upfront use of NVIDIA GPUs and need for speed, the GPU-accelerated NVIDIA Parabricks genomic analysis suite was a natural fit for secondary analysis. Another important factor was the fact that Parabricks supports DeepVariant, where GPU acceleration yields a FASTQ-to-VCF turnaround time of around 16 minutes for 30x human whole genome sequencing experiment.
One of the greatest strengths of DeepVariant is its flexibility – anyone can train the DeepVariant model to maximize performance for a given sequencing system or application. The Singular R&D team regularly trains and tests new DeepVariant models to improve potential optimizations to sequencing and base calling. Additionally, regular retraining of existing public facing models to produce additional reference datasets from whole genome or targeted sequencing of NIST reference materials is performed. This allows Singular to incrementally improve model accuracy.
For Singular Genomics, a single training event will often entail parallel testing of multiple DeepVariant models given that multiple model parameters may be customized by the user. This includes interesting new channels such as insert-size, GC content, and custom-defined read or base-level tags that may improve performance. We also find it useful to examine parameters controlling the example generation process, such as the minimum MAPQ and allele frequency threshold.
The computational lift required to train a model is dependent on whether you would like to produce a “warm start” model trained from a previous model, or rather build a new model from scratch through “de novo” training. Although warm start training may be the most appropriate choice for incrementally improving an existing model (say, by addition of new reference data), de novo building may be required to test the impact of certain non-standard parameters such as the custom tags mentioned above. De novo model building requires a much heavier computational lift owing to the need to train from a much larger dataset, making any acceleration of the training process especially impactful.
Whether using a warm start or a de novo approach, training a new DeepVariant model consists of a `make_examples` step, where the examples of variants in the aligned BAM data is converted into a tensor format, followed by repeated cycles of training and testing. As a final step, the trained model is validated, typically by variant calling and analysis of a NIST reference genome that was not used in training. While the variant calling portion of the workflow had already been accelerated in Parabricks, the `make_examples` step has until now required a lengthy CPU-driven process. The new update removes this bottleneck, allowing for faster and cheaper model training.
Singular Genomics benchmarked the acceleration of DeepVariant in Parabricks by examining the time required to conduct warm start training of the default DeepVariant v1.4 Illumina WGS model using a single 30x WGS dataset derived from sequencing of the NIST sample NA12878 (HG001) on the G4 platform, training on chromosomes 1-19, and testing on chromosome 21. The workflow is implemented on the Broad Institute of MIT and Harvard’s Terra cloud platform, where the focus on workflow transparency and ease of development make it a good fit for this type of task.
In September 2022, an NVIDIA Parabricks workspace with six pre-configured workflows was made available on Terra for its over 25,000 researchers to quickly analyze massive amounts of healthcare data. Parabricks workflows in Terra deliver accelerations of up to 24x faster execution compared to equivalent CPU-based workflows, and cut the total cost of execution by up to 50%. Terra provides an easy to use platform for running and sharing pre-loaded and pre-configured workspaces, so researchers should have no issue creating their own copies of the Parabricks workspace if they want to try it out for themselves.
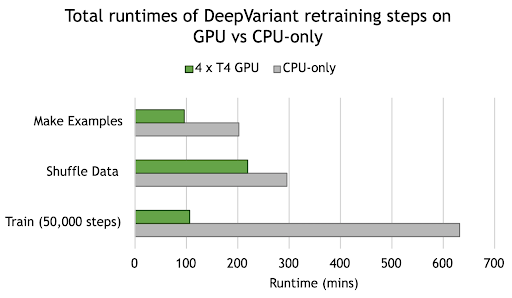

Figure 2. NVIDIA GPUs provide acceleration across all stages of DeepVariant Retraining. Runtime for each stage is the total for running on chr1-19 for training and chr21 for testing. Compute instances selected for benchmarking included 240GB memory, 64 x vCPU, and 4 x T4 GPU.
Using the DeepVariant retraining tool in Parabricks, Singular Genomics observed 2.1x acceleration in `make_examples`, from 3.4 hours on CPU-only to 1.6 hours on 4 NVIDIA T4 GPUs. We also see a 5.9x acceleration in model training, from 10.5 hours on CPU-only to 1.8 hours on GPU. These, and other improvements, translate to a total time saving of just under 12 hours across the entire retraining process, from approximately 19 hours down to 7 hours turnaround time.
Thus, this is appears to be a very nice improvement to the training process.
This reduced runtime has an impressive result on overall costs. The underlying CPU instance runs at a cost of $4.384/hour, with each additional T4 GPU adding $0.35/hour, resulting in a total of $5.784/hour for the GPU instance benchmarked here. As such, the cost of retraining on CPU-only, with a runtime of 19 hours, totaled $82.67, whereas with the additional 4 T4 GPUs and a runtime of 7 hours this totaled $40.84, a cost saving of just over 50%.
Finally, to confirm that the training worked, Singular tested their newly trained DeepVariant model with G4 data by performing germline whole genome analysis of NIST HG002 sequenced to ~30x coverage. The table below shows results from hap.py analysis of high confidence regions for the default DeepVariant v1.4 Illumina WGS model and the newly trained model. Significant improvements in the indel F1 score were observed, indicating that the training was successful.
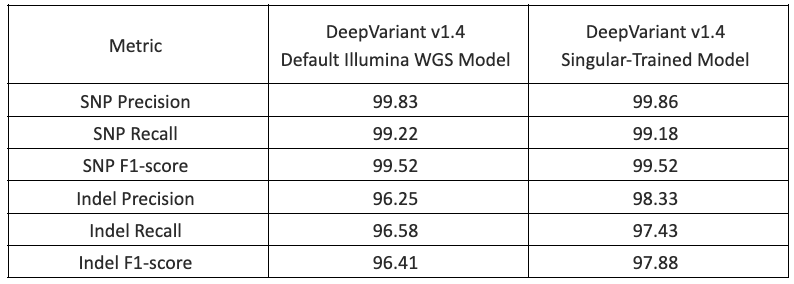

Table 1. Assessment of the training framework accuracy by Singular on Singular GIAB data, showing improved Indel calling (Indel F1-score) and equivalent SNP calling (SNP F1-score).
In summary, the DeepVariant retraining tool was very easy to use and helped Singular Genomics fine tune DeepVariant to their instrument output to improve variant calling. Singular Genomics is excited to make regular use of this new Parabricks retraining tool to improve variant calling. Parabricks DeepVariant retraining tool is now available on Terra for everyone to fine tune DeepVariant on your own data and improve variant calling.
Try it out for yourself today
The DeepVariant retraining tool in the newest release of Parabricks 4.1 is available on Terra for anyone to retrain DeepVariant for more accurate variant calling. The NVIDIA Parabricks Terra workspace was created by the NVIDIA team and is preloaded with example data, workflow configurations, and straightforward instructions, so you can try out the workflows without having to install or tweak anything. Simply clone the workspace and launch the preconfigured examples or load your own data to get started.
If you’re new to running workflows on Terra, see the Workflows Quickstart Tutorial.
Don’t hesitate to reach out if you have any questions or if you run into any trouble running the workflows. For help with Terra-specific features (e.g. how to launch, monitor and troubleshoot WDL workflows in Terra), you can either post in our public discussion forum or contact the helpdesk team privately. For technical questions about NVIDIA Clara Parabricks, please visit the developer forum page here.








