The past few years have seen a massive surge in the development of advanced analytical methods for biomedical research, fueled in part by technological innovations that allow computational scientists to crunch data at ever-increasing speed and scale. A growing number of technology companies have joined the effort to help researchers tackle emerging challenges, ranging from large-scale genomics to multi-modal analysis of the myriad data types associated with medical records — including doctors’ notes, which are famously easy to read and interpret.
Today NVIDIA, a pioneer in AI and accelerated computing, announced a new partnership with the Broad Institute that will pool the two organizations’ respective expertise in deep learning, accelerated compute, and biomedical research. This partnership builds on an existing collaboration between NVIDIA and the Broad’s GATK team, who have already been working together to improve some of the deep learning algorithms in GATK. (Keep an eye on the GATK blog for an upcoming release announcement.)
The NVIDIA team released a Clara Parabricks workspace in Terra that makes their GPU-accelerated genomic analysis toolkit available on the cloud at the click of a button. As shown by the benchmarking results below, the Clara Parabricks workflows in Terra deliver accelerations up to 24x faster execution compared to equivalent CPU-based workflows, and can cut the total cost of execution by up to 50%.
What’s in the box? Drop-in replacements for popular GATK workflows
NVIDIA Clara Parabricks is a suite of GPU-accelerated industry-standard tools for the most common genomics analyses, including read alignment and both germline and somatic variant calling for whole genomes, exomes, and gene panels.
To make these tools easy to run in Terra, the NVIDIA team produced six modular workflows written in the Workflow Description Language (WDL) that are designed as drop-in replacements for the corresponding GATK workflows, summarized in the figure below.
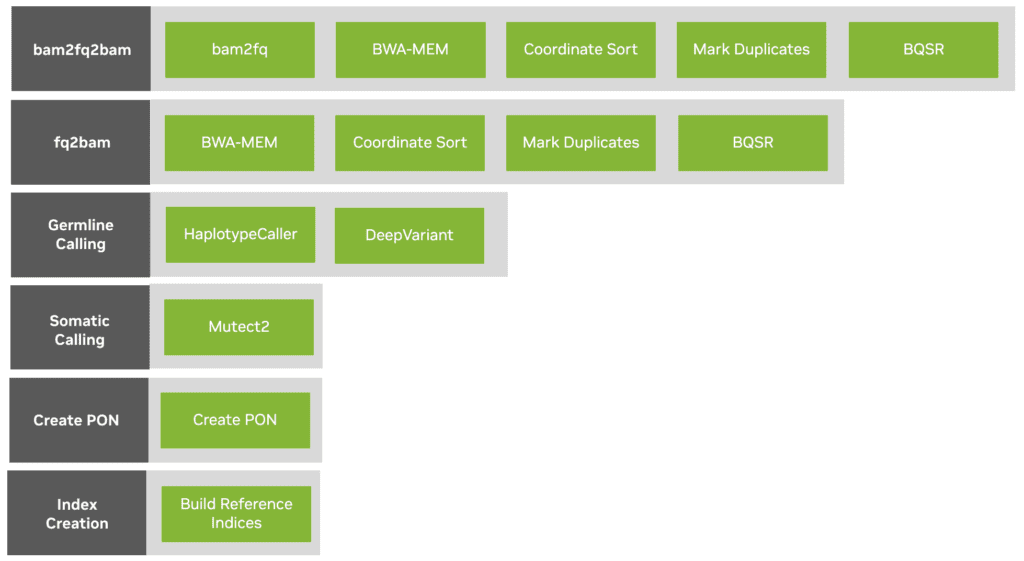

The six Clara Parabricks workflows available as WDLs in Terra (leftmost boxes), with component modules listed to their right. In the case of the germline calling workflow, the two modules (HaplotypeCaller and DeepVariant) are alternative options that can be toggled with a configuration flag.
Each workflow comes with a reference configuration that includes the most appropriate GPU instances to run them on, and the ability to select GATK best practice flags and options.
The NVIDIA team collaborated with the GATK team at the Broad Institute to evaluate the accuracy of the germline workflows. Through this rigorous process, they verified that the Clara Parabricks workflows produce results that are functionally equivalent to the CPU-native GATK versions, as originally defined here.
As a specific example, benchmarking on publicly available Genome in a Bottle (GIAB) samples with the fq2bam and germline caller workflows from the Clara Parabricks suite produced variant calling results that were >0.9999 equivalent in both precision and recall to those produced by the BWA, MarkDuplicates, BQSR, and HaplotypeCaller commands in the GATK’s Whole Genome Germline Single Sample variant calling workflow (available here in Terra).
Up to 24 times the speed and half the cost
The team benchmarked the runtime of the Clara Parabricks workflows on Terra, and found that the GPU-accelerated workflows delivered speedups of up to 24x for germline genome analyses.
When using Clara Parabricks in Terra, the total runtime on NVIDIA GPUs is reduced significantly for a 30x whole genome including BWA-MEM, MarkDuplicates, BQSR, and HaplotypeCaller. Total run time from FASTQ to VCF including variant calling with haplotypecaller, is just over 2 hours on NVIDIA T4 GPUs compared to 24 hours in a CPU-based environment. Additionally, the cost for analysis was 50% reduced in the GPU environment compared to the CPU environment.
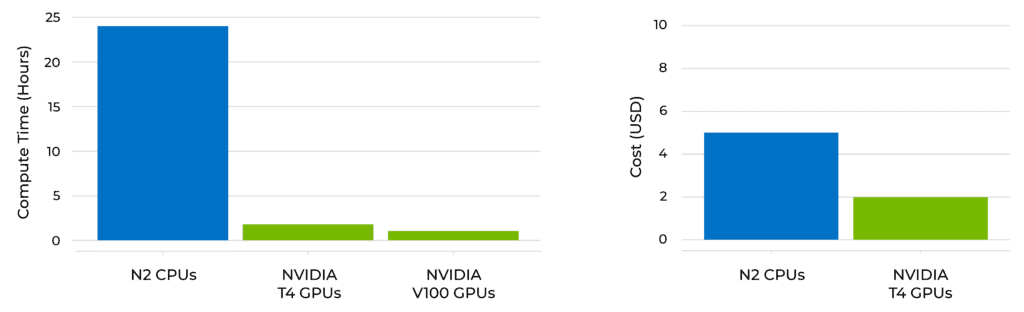

Time and cost comparisons of alignment and variant calling (FastQ to VCF) on CPU vs. GPU for a 30X whole genome in Terra.
Runtime from FASTQ to BAM (with BWA-MEM) was reduced from 7 hours with CPU instances (N2) to a little over an hour with 4 NVIDIA T4 instances, and dropped even further to ~45 mins with 8 NVIDIA V100 GPU instances.
You can also see in the figure above (right side panel) that the overall cost of running the workflows on the T4 GPUs is less than half the cost of running the CPU-based equivalents, which is always a happy surprise.
I’ve written before about the speed benefits of GPUs on this blog, though that was in the context of interactive analysis. One of my big takeaways was that you have to find the sweet spot between speed and cost that works for you, because oftentimes the fancy hardware that makes things go really fast is also the most expensive to rent. The good news is that if the speedup is big enough, you only have to use the special hardware for a very short amount of time, and that makes up for the higher rate. Or in the case of the T4 instances, more than makes up for it, since that configuration manages to be substantially more economical even as it delivers a heck of a speed boost.
According to NVIDIA, the T4 GPU is designed to optimize cost and performance when running inference-heavy workloads like Clara Parabricks — which matches what we’re seeing in these benchmarking results. NVIDIA T4 GPUs are available for as little as $0.11 per hour on Terra (backed by Google Cloud), so by running the Clara Parabricks workflows there, you can run the entire alignment and variant calling pipeline for less than $2.50 a sample. That represents a major reduction from the $5 cost-per-sample of the GATK’s CPU-based workflows (running on N2 instances) while still reducing the overall data processing runtime from a whole day to less than three hours. Instances on Terra can also be configured with up to 8 V100 GPUs, which are more optimized for absolute performance than the T4. The same fastq-to-VCF pipeline on 8 V100 GPUs is up to 24X faster than the CPU pipeline, at roughly twice the cost.
I’m excited to see this technology being made available to a wide audience, in a form that doesn’t require taking specialized training or purchasing expensive hardware. It’s a big step forward toward ensuring researchers of any background are able to run sophisticated genomic analysis at scale.
Try it out for yourself today
The Clara Parabricks Terra workspace created by the NVIDIA team is preloaded with example data, workflow configurations, and straightforward instructions, so you can try out the workflows without having to install or tweak anything. Simply clone the workspace and launch the preconfigured examples, or load your own data and get to work.
If you’re new to running workflows on Terra, see the Workflows Quickstart Tutorial.
Don’t hesitate to reach out if you have any questions or if you run into any trouble running the workflows. For help with Terra-specific features (e.g. how to launch, monitor and troubleshoot WDL workflows in Terra), you can either post in our public discussion forum or contact the helpdesk team privately. For technical questions about NVIDIA Clara Parabricks, please visit the developer forum page here.
Acknowledgments
We are grateful to the NVIDIA team, specifically Eric Dawson and Vanessa Braunstein, for running the workflow benchmarks and helping us characterize the benefits of using Clara Parabricks on GPU instances in Terra.
Resources
– Terra Workflows Quickstart Tutorial
– Terra blog about using GPUs for interactive analysis, machine learning
– Cromwell documentation about runtime parameters for using GPUs in workflows
– Clara Parabricks Genomic Analysis webpage
– Clara Parabricks Documentation Page
– Clara Parabricks in Terra Workspace
– Clara Parabricks GTC DLI Hands on free workshop on Sept 21 as part of GTC



