Laury Mignon is Executive Director of Translational Medicine at Ionis Pharmaceuticals. She is responsible for improving the probability of (technical) success of Ionis’ preclinical assets from “bench to bedside”. In this guest blog post, Laury presents a novel approach to study participants’ data access and control in industry-sponsored clinical studies.
Whole-genome sequencing (WGS) is increasingly used in human research, including clinical trials, and the resulting data hold a lot of potential value for research beyond the immediate use for which it is collected. Allowing pharmaceutical companies to use WGS data from clinical trials for exploratory research could unlock substantial benefits for patients as well as for study participants who could receive actionable incidental findings. However, this raises important questions around participant consent, disclosure of incidental findings, and the rights of participants to withdraw their data from further study.
To address these questions, we developed the Exploratory Genetic Research Project (EGRP), a novel framework that aims to provide an umbrella protocol to collect genetic material in all of a sponsor’s clinical studies, giving consenting individuals the right to access and control access to their genetic data while enabling unspecified or exploratory future research. We recently published a manuscript describing the full EGRP protocol, as well as the detailed reasoning behind key design decisions we made, and we are currently working with Color Genomics and the Broad Institute to test our very first implementation of this protocol.
By Laurence Mignon, Kim Doan, Michael Murphy, Lauren Elder, Chris Yun, Jeff Milton, Shruti Sasaki, Christopher E. Hart, Dante Montenegro, Nickolas Allen, Dany Matar, Danielle Ciofani, Frank Rigo, and Leonardo Sahelijo (2022)
In Contemporary Clinical Trials, Volume 119, https://doi.org/10.1016/j.cct.2022.106819
I hope you will read the paper to get the full picture of this innovative framework. Here, I wanted to highlight our decision to use a secure cloud platform — specifically Terra — to store the WGS data and make it available for analysis in a way that protects the privacy and autonomy of study participants.
Terra as the cloud-based Sandbox
Our intention was to prioritize participants’ control over the use of their genetic data, while enabling proprietary future research on the clinical and genetic data. For that reason, the EGRP process stipulates that the pharmaceutical company who sponsors the study is not permitted to download participants’ individual WGS data onto their own servers. Instead, the WGS data must remain in the custody of an independent partner, the data host, who performs the genome sequencing and makes the data available for analysis through a secure cloud platform.
Additionally, samples are de-identified prior to sequencing through the use of barcodes managed by a third partner, the “honest broker”, who is responsible for interfacing with participants and managing consents, barcodes and return of results.
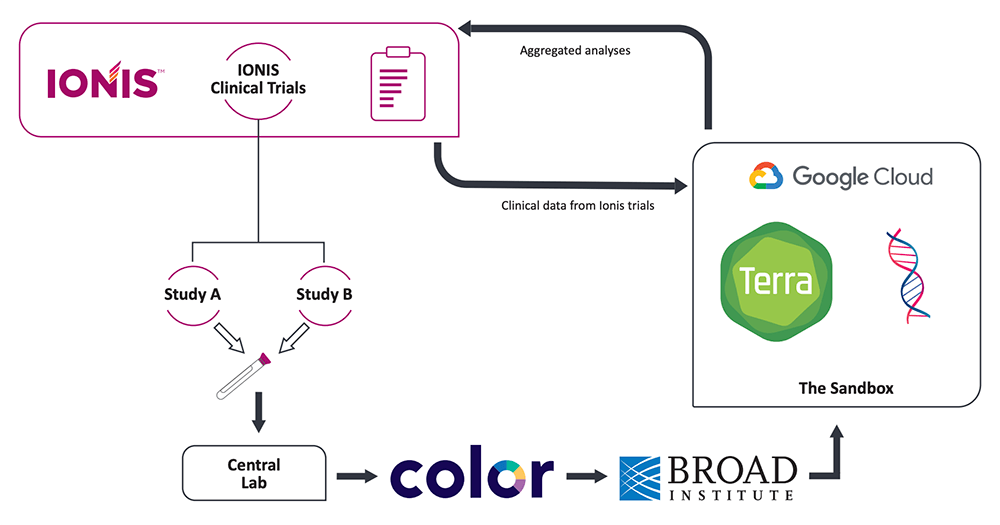

Flow of information between the EGRP partners, with Ionis as the sponsor (pharmaceutical company running clinical trials), Color Genomics as the honest broker (interfacing with participants, managing consents, barcodes and return of results) and the Broad Institute as the data host (performing WGS and providing secure access to data in Terra). Modified from Mignon et al., 2022.
This setup makes it possible to ensure that data can be promptly removed from the system if a participant decides to withdraw their data. All the participant needs to do is notify the honest broker (Color Genomics), who will then issue a withdrawal order for the corresponding barcode to the data host (Broad Institute). By excluding the sponsor from the removal process, we effectively remove potential conflicts of interest and increase the amount of control that study participants can wield over their genetic data.
In addition, using Terra offers the opportunity to use a large set of cloud computing tools that are readily available in the platform and in many cases, have been optimized for genomic analysis at scale. This includes algorithms and pipelines created by other Terra users, data engineers supporting large projects such as the All of Us Research Program and the Human Cell Atlas, and the wider bioinformatics community. We view this as a promising path toward a faster and more standardized way of performing genetic analyses, as well as a fairer method of developing and sharing computational tools across private and public industries.
As we move forward with our first real-world test of the EGRP protocol, we are excited to have defined a process that serves the interests of individual study participants and pharmaceutical sponsors, and we are hopeful that it will provide a blueprint for future work by other clinical trial sponsors as well.



