Cancer researchers are increasingly turning to cloud resources, primarily to access data and solve large-scale computational problems. Yet as the infrastructure matures, the cloud can also be used to widen access to important tools to clinicians and point-of-care personnel who do not have the level of computational training that has traditionally been required to use such tools.
In this guest blog post, Brendan Reardon, computational biologist in the Van Allen Lab at Dana Farber Cancer Institute, tells us about the MOAlmanac algorithm and the analysis portal that he and his collaborators built on top of Terra to enable a wider range of users to leverage large-scale genomics to guide individualized patient care in oncology.
In recent years, it has become routine to use individual tumor molecular profiling to identify “first-order” genomic alterations such as single-gene variants that can inform therapeutic options. However, clinical interpretation algorithms and knowledge bases do not generally account for the interactions between these first-order events and global molecular features such as mutational signatures, despite the increasing recognition that these “second-order” events can be associated with clinical outcomes.
To address this gap, our team developed the Molecular Oncology Almanac (MOAlmanac), an open-source clinical interpretation algorithm paired with a novel knowledge base to enable integrative interpretation of genomic and transcriptional cancer data.
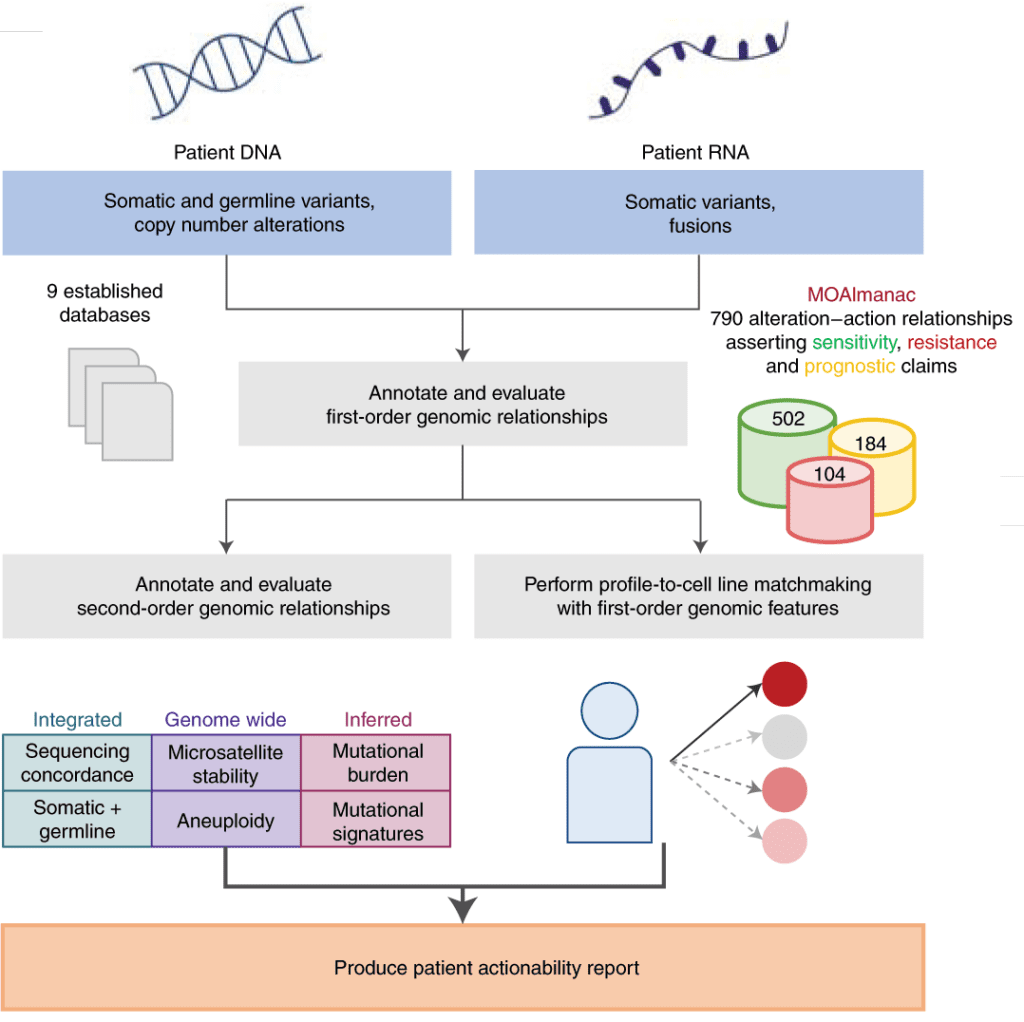

Overview of the MOAlmanac method (from Reardon et al, 2021, Figure 1a)
In our recent manuscript published in Nature Cancer, we showed how the MOAlmanac method nominated a median of two therapies per patient and identified therapeutic strategies administered in 46% of patient profiles when applied to a prospective precision oncology trial cohort. These results suggest MOAlmanac can be readily applied for translational hypothesis generation in cancer research, yet the ultimate goal of the project is to be able to also assist clinicians in their point-of-care treatment decision-making.
To that end, we are now working with other groups to test the method in real-world projects. In a collaboration with Dr. Katherine Janeway and Dr. Nikhil Wagle of Dana-Farber Cancer Institute, we are applying the MOAlmanac approach to participant data from the Count Me In Osteosarcoma Project as part of the National Cancer Institute’s Participant Engagement and Cancer Genome Sequencing (PE-CGS) Network. The collaboration aims to evaluate the use of MOAlmanac as an aid to assist molecular pathologists in the clinical interpretation and sample contextualization for somatic return of results to study participants.
As a rare type of cancer that is severely understudied, osteosarcomas pose a particular set of challenges and are in dire need of innovative approaches to drive novel treatment strategies, clinical trials, and standards of care. While MOAlmanac is currently released for research use only, we believe it constitutes an exciting step forward toward the next level of precision cancer medicine.
Using the cloud to widen access to computational oncology
In order to ensure that this promising method is widely accessible and usable by researchers and clinicians who do not have extensive computing experience, we created a website that enables others to easily browse the MOAlmanac knowledge base and run the MOAlmanac algorithm on their own samples.
The MOAlmanac portal interface walks users with minimal computational experience through the basic steps of setting up the analysis; then it uses Terra’s built-in data modeling and workflow management capabilities under the hood to run the analysis efficiently and securely. This enables users such as point-of-care clinicians to take advantage of Terra’s powerful and highly scalable workflow execution system without having to learn how to use Terra itself.
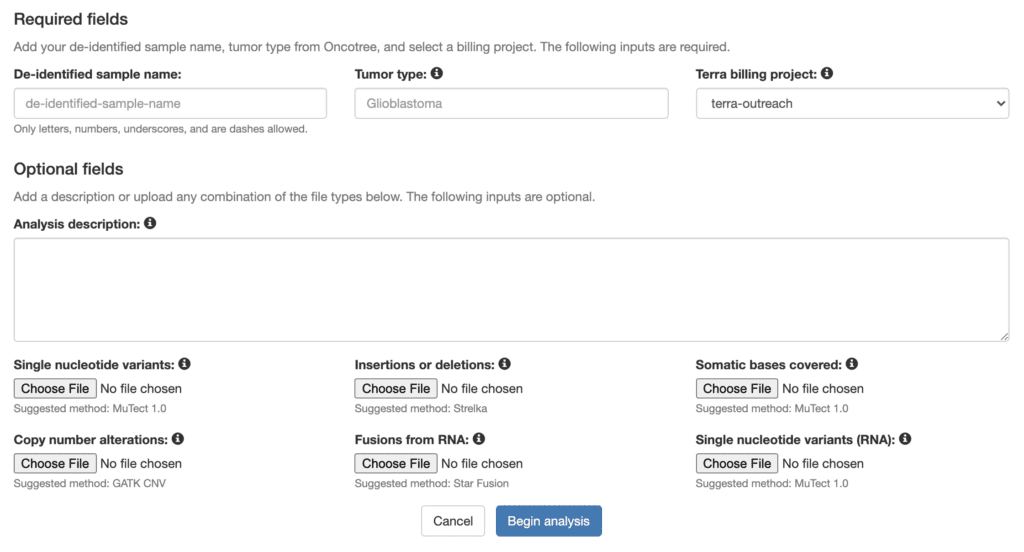

Screenshot of the analysis configuration page on the MOAlmanac portal
For researchers who are comfortable with bioinformatics and are interested in running the MOAlmanac method on their own infrastructure, customizing it or extending it with their own ideas, we provide complementary resources, including source code, containerized tools and scripted workflows. We also made a public Terra workspace that demonstrates the use of these resources; we invite you to try it out and let us know what you think by emailing us at moalmanac[at]ds.dfci.harvard.edu or tweeting at us, @moalmanac.



