The Broad Institute has been an epicenter of activity in terms of COVID-19 response, taking on a significant chunk of COVID-19 testing locally in MA, as well as an active role in multiple research projects and public health initiatives. As a result, many of our colleagues in the Broad’s Data Sciences Platform (DSP) have been called on to step up, either individually or with their entire teams, to support these efforts. We’ve previously featured some of those efforts in blog posts, videos, and resource pages. In this guest blog, Andrew Zimmer, Director of the Data Donation Platform within the Broad Institute Data Sciences Platform, recounts how his team came to support TestBoston, a collaborative project with Brigham and Women’s Hospital that aims to monitor the infection rate of COVID-19 in Massachusetts over time.
Early in the pandemic, three physician principal investigators in the wider Broad community, Drs. Ann Woolley, Lisa Cosimi, and Deborah Hung, realized the need for a large-scale, unbiased study that would allow us to understand some really fundamental metrics about COVID-19 in our community, and answer some basic questions about this virus that was new to the entire world. Their plan, which would eventually become the largest longitudinal study of its type in the US, called for a robust and scalable way to recruit participants, track survey data, coordinate the logistics of shipping and receiving test kits, and share data securely with researchers.
That was right up my team’s alley, so we jumped at the chance to help support COVID-19 response efforts. Fast forward a year into the pandemic, and to date, our work has helped enroll over 10,000 participants over 3 months in the TestBoston research study and continues to support sample collection and data sharing for this ongoing research project.
This is the story of how we made it work.
Partnering with the population to keep tabs on COVID
One of the challenges of this pandemic has been the difficulty of getting a clear picture of how far the virus has spread through our communities, in part because of how differently it affects different people. We know the virus can cause very serious, even fatal disease; yet some people who catch it experience only mild symptoms. Importantly, now that we know that many infected people can be completely asymptomatic. If we only test people who experience symptoms or who know they have been in contact with someone who has tested positive, we only get a partial, highly fragmented picture of the true situation. The basic idea of the TestBoston study is that we can get a more complete picture by testing a relatively more unbiased set of volunteers in the community on a regular basis, whether they feel sick or not.
In practice, this involves collecting nose swabs and finger-prick blood samples from participants at regular intervals, then testing these for current infection (by PCR) and past infection (by antibody testing). Given the circumstances, the study was designed to run entirely remotely: participants are invited to sign up online (which involves a few survey questions and a consent form), receive sample collection kits in the mail, and send the samples back to the lab using prepaid return kits. This minimizes the logistical burdens placed on participants and the risk of viral spread that in-person interactions would entail, and makes it possible to return results directly to participants via an online dashboard.
From the point of view of the participants, the process is very straightforward, but the simplicity and ease-of-use of the TestBoston.org website belies the underlying complexity of the project.
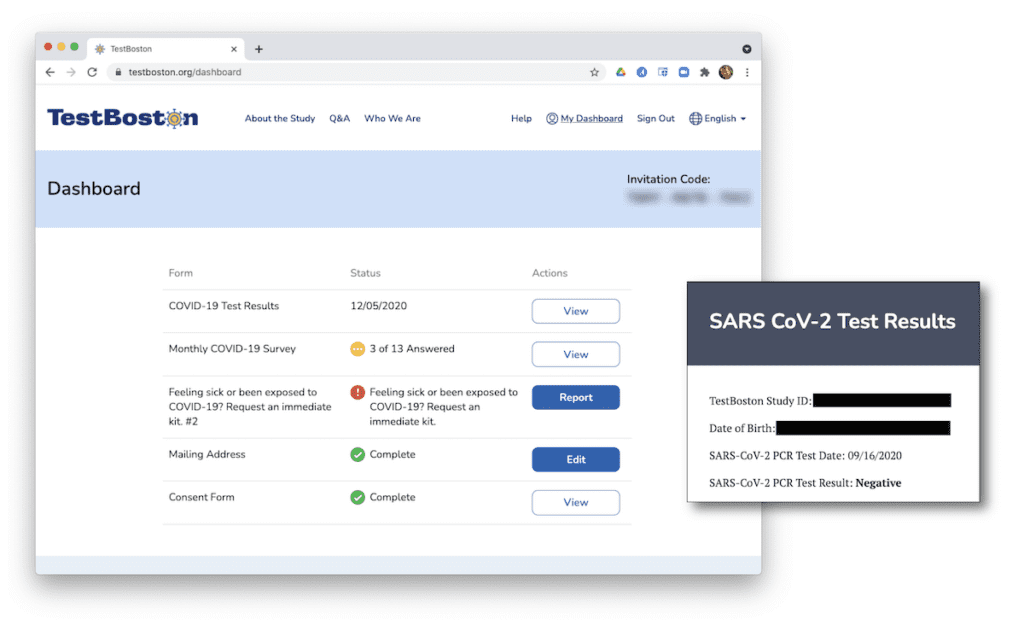

Behind the scenes, there are many moving parts involved in building and operating the computing infrastructure to support such an ambitious study. There are technical and logistical considerations, like integrating with shipping companies and sharing data securely with study staff. There are also some complex regulatory and procedural requirements associated with human subjects research, collaborations with hospitals, and an Emergency Use Authorization granted by the FDA that allowed us to give study participants the results of their COVID-19 tests.
Building a platform for such an endeavor from scratch could easily take over a year’s worth of work. I’m proud to say that our team delivered a functional platform in just four months, while simultaneously helping scale up the Broad’s COVID-19 testing infrastructure during the early days of the pandemic.
How did we accomplish this? It just so happens that we had already built an open-source software framework, called Pepper, that met many of TestBoston’s requirements (including the complete absence of in-person clinic visits) and that we were able to adapt and deploy on fairly short notice.
Introducing Pepper, an open-source software framework for patient-partnered research
Pepper was born out of a deep collaboration with Count Me In, a nonprofit organization committed to advancing patient-partnered cancer research. If you’re not familiar with the concept, patient-partnered research aims to connect researchers directly with patients and advocacy groups, to break down traditional silos, and increase the utility of patient data. As described on their website, Count Me In “enables interested patients to share their saliva, blood, stored tumor samples, clinical information, and experiences to help researchers detect new and important patterns in cancer progression and response to treatment across large numbers of people.”
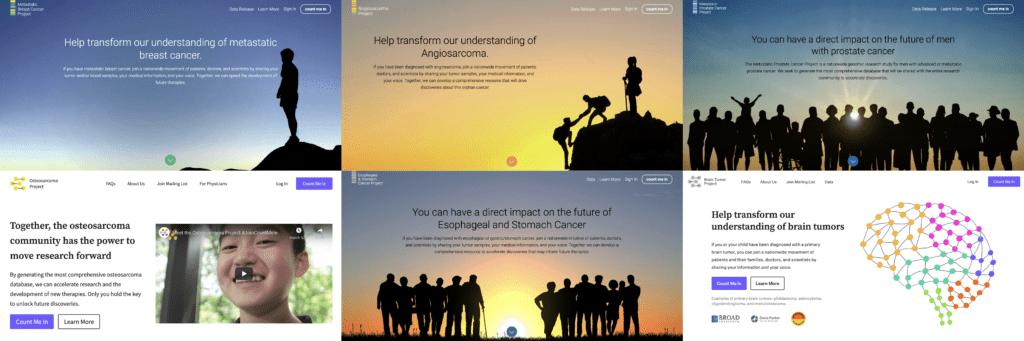

The six patient-partnered research projects currently supported by Count Me In: the Metastatic Breast Cancer Project, the Angiosarcoma Project, the Metastatic Prostate Cancer Project, the Osteosarcoma Project, the Esophageal and Stomach Cancer Project, and the Brain Tumor Project.
We originally started working with Count Me In as part of a new initiative to help scale patient-partnered research projects through online technologies. We began by building a few standard websites to enable prospective participants to sign up for cutting-edge research projects, a process that typically revolves around securing the participant’s informed consent. On a project-by-project basis, this was fairly straightforward, but as the number of projects grew from a handful to dozens, it became clear that copy-pasting web software wasn’t going to scale well.
Ultimately, we realized what we really needed was a proper multi-tenant platform that would be capable of hosting hundreds of studies amounting to thousands of patients, streamlining much of the book-keeping operations of at-home sample and data collection directly from participants.
So we built one. We didn’t completely reinvent the wheel, of course — we leveraged numerous existing “software-as-a-service” offerings that provided key pieces of functionality, and focused most of our de novo development on building a framework that would tie all these pieces together. We are also creating software development kits (SDKs) and administrative modules that will enable study managers to deploy and customize their project websites by themselves, to maximize their freedom and minimize the amount of intervention that would be required from our team.
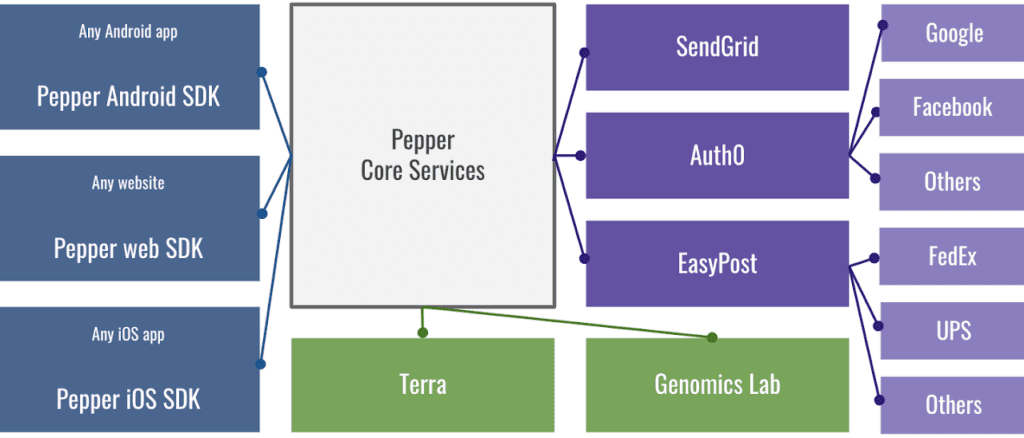

Modular services that plug into the “core” of Pepper: (1) Blue/left: SDKs for making websites and mobile apps; (2) Green/bottom: integration with the Broad Genomics Lab (for running tests) and with Terra for data sharing; (3) Purple/right: service integrations including SendGrid email service, Auth0 authentication for registration/login, EasyPost for shipping services.
In that same spirit of empowering project administrators, we designed the Pepper framework to run on the cloud, which eliminates any dependence on local infrastructure and makes it possible to reliably deploy projects using cookie-cutter recipes that automatically marshall computing resources and set everything up just right.
This is also something that could be daunting to undertake from scratch, but thankfully we had a lot of in-house cloud engineering experience to draw on. Our colleagues in the Data Sciences Platform had been building and operating cloud services for some time, including the Broad’s data processing infrastructure and, of course, the Terra platform. In fact, we are actively working on integrating Pepper with Terra for storing and sharing study data with authorized researchers.
Through several iterations of the Pepper framework, we’ve been able to drive down the costs involved in design, engineering, and deployment of individual projects by about a factor of 5, and we are continuing to work to optimize the framework in order to minimize costs and maximize the impact of every research dollar.
Pepper now supports six major cancer research projects under the aegis of Count Me In, as well as a growing number of projects spanning a range of disease domains, from ataxia telangiectasia to prion disease. And of course, the TestBoston project!
Chance favors the prepared mind
This quote from Louis Pasteur, which has resonated with me ever since I first heard it from my 10th-grade biology teacher, is an apt summary of how the engineering came together for TestBoston. A key part of the Broad’s philosophy is to invest not just in specific scientific projects, but also in infrastructure and technology platforms that make it possible to move rapidly to innovate, leveraging and extending capabilities from one project to another. Our team’s department in particular, the Broad’s Data Sciences Platform, was created five years ago for the explicit purpose of building tools and infrastructure to meet the emerging data science needs of the research community.
So, while we didn’t expect COVID-19 itself, we were in many ways quite well prepared to support response efforts, having just spent several years developing both a robust engineering organization and an array of highly relevant software products, including Pepper and Terra, in pursuit of our mission to support data science. We were able to lean on the deep network of scientific collaborations and industry partnerships that we had formed in the process; including our long-time partner, Intel, who provided funding through their PRTI fund* to operate the TestBoston platform.
We were also lucky in a few places that matter. This is complicated and challenging work, and with so many moving parts, we can never take success for granted.
Yet that makes it all the more gratifying when we see our labor bear fruit, particularly in an application like this where lives are literally at stake. For many of our team members whose expertise lies in domains other than medicine or bench science — software engineers, designers, project managers, and more — the chance to use their skills to help fight this pandemic in a tangible way has been a great source of pride and comfort.
Resources
Pepper is an open-source project developed by the Broad Institute of MIT and Harvard, released under a BSD-3 clause license. The source code is distributed across several component repositories listed here.
TestBoston is a COVID-19 study supported by Brigham and Women’s Hospital and the Broad Institute of MIT and Harvard.
*Intel Funding for this was provided in part by Intel Corporation’s Pandemic Response Technology Initiative. Intel is committed to creating a more responsible, inclusive, and sustainable world enabled through technology and collective actions.
Count Me In is a nonprofit organization committed to advancing patient-partnered research, stewarded by Emerson Collective, a California-based social change organization; the Broad Institute of MIT and Harvard; the Biden Cancer Initiative, an independent nonprofit organization that builds on the federal government’s Cancer Moonshot; and the Dana-Farber Cancer Institute, a leading cancer hospital.



